Modified pegRNAs mitigate scaffold-derived prime editing by-products
- PMID: 40204736
- PMCID: PMC11982355
- DOI: 10.1038/s41467-025-58653-1
Modified pegRNAs mitigate scaffold-derived prime editing by-products
Abstract
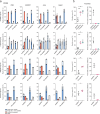
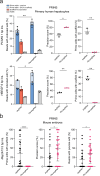
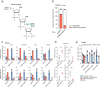
Prime editors (PEs) employ reverse transcriptase (RT) to install genomic edits using a template within the prime editing guide RNA (pegRNA). RT creates a 3' genomic flap containing the intended edit. However, reverse transcription can continue beyond the template, incorporating the pegRNA scaffold sequence into the 3' flap. These scaffold-derived by-products can be installed alongside the intended edit, reducing prime editing precision. Here, we develop a method that prevents RT from accessing the scaffold, thereby mitigating such by-products. We demonstrate that an internal abasic spacer or 2'-O-methylation within the pegRNAs terminates RT at the end of the template. This prevents scaffold-derived sequences from being incorporated into the target locus. We benchmark these pegRNAs in different cell types and demonstrate that they can be used with processive PEs such as PE6d or PE**. Our findings provide a simple approach to mitigate a common prime editing by-product and improve prime editing precision.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.A., L.D., H.M., K.M.B., A.L.L., A.S., E.G., S.M., G.T., P.P.H., S.Š., M.F., N.A., M.M., M.P are employees of AstraZeneca and may be AstraZeneca shareholders. L.D. and A.S. are funded by Promega corporation. AstraZeneca filed patents related to this work (WO2021204877A2 and WO2023052508A2).
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

