Proteomics and personalized PDX models identify treatment for a progressive malignancy within an actionable timeframe
- PMID: 40204966
- PMCID: PMC11982353
- DOI: 10.1038/s44321-025-00212-8
Proteomics and personalized PDX models identify treatment for a progressive malignancy within an actionable timeframe
Abstract
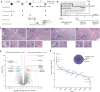
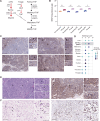
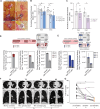
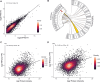
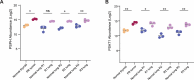
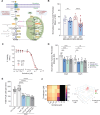
Genomics has transformed the diagnostic landscape of pediatric malignancies by identifying and integrating actionable features that refine diagnosis, classification, and treatment. Yet, translating precision oncology data into effective therapies for hard-to-cure childhood, adolescent, and young adult malignancies remains a significant challenge. We present the case for combining proteomics with patient-derived xenograft models to identify personalized treatment for an adolescent with primary and metastatic spindle epithelial tumor with thymus-like elements (SETTLE). Within two weeks of biopsy, proteomics identified elevated SHMT2 as a target for therapy with the anti-depressant sertraline. Drug response was confirmed within two months using a personalized chicken chorioallantoic membrane model of the patient's SETTLE tumor. Following failure of cytotoxic chemotherapy and second-line therapy, the patient received sertraline treatment and showed decreased tumor growth rates, albeit with clinically progressive disease. We demonstrate that proteomics and fast-track xenograft models provide supportive pre-clinical data in a clinically meaningful timeframe to impact clinical practice. By this, we show that proteome-guided and functional precision oncology are feasible and valuable complements to the current genome-driven precision oncology practices.
Keywords: Genomics; Patient Derived Xenografts; Pediatric Cancer; Precision Therapeutics; Proteomics.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures
Similar articles
-
Integration of genomics, high throughput drug screening, and personalized xenograft models as a novel precision medicine paradigm for high risk pediatric cancer.Cancer Biol Ther. 2018;19(12):1078-1087. doi: 10.1080/15384047.2018.1491498. Epub 2018 Oct 9. Cancer Biol Ther. 2018. PMID: 30299205 Free PMC article.
-
Patient-derived xenograft (PDX) models of colorectal carcinoma (CRC) as a platform for chemosensitivity and biomarker analysis in personalized medicine.Neoplasia. 2021 Jan;23(1):21-35. doi: 10.1016/j.neo.2020.11.005. Epub 2020 Nov 16. Neoplasia. 2021. PMID: 33212364 Free PMC article.
-
Utilizing Patient-Derived Xenografts to Model Precision Oncology for Biliary Tract Cancer.Clin Cancer Res. 2025 Jan 17;31(2):387-402. doi: 10.1158/1078-0432.CCR-24-1233. Clin Cancer Res. 2025. PMID: 39513959 Free PMC article.
-
Pancreas Cancer Precision Treatment Using Avatar Mice from a Bioinformatics Perspective.Public Health Genomics. 2017;20(2):81-91. doi: 10.1159/000479812. Epub 2017 Sep 1. Public Health Genomics. 2017. PMID: 28858862 Review.
-
Generation and application of patient-derived xenograft models in pancreatic cancer research.Chin Med J (Engl). 2019 Nov 20;132(22):2729-2736. doi: 10.1097/CM9.0000000000000524. Chin Med J (Engl). 2019. PMID: 31725451 Free PMC article. Review.
References
-
- Akhoundova D, Rubin MA (2022) Clinical application of advanced multi-omics tumor profiling: Shaping precision oncology of the future. Cancer Cell 40:920–938 - PubMed
-
- Berlanga P, Pierron G, Lacroix L, Chicard M, Adam de Beaumais T, Marchais A, Harttrampf AC, Iddir Y, Larive A, Soriano Fernandez A et al (2022) The European MAPPYACTS trial: precision medicine program in pediatric and adolescent patients with recurrent malignancies. Cancer Discov 12:1266–1281 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

