Translational genomics of osteoarthritis in 1,962,069 individuals
- PMID: 40205036
- PMCID: PMC12119359
- DOI: 10.1038/s41586-025-08771-z
Translational genomics of osteoarthritis in 1,962,069 individuals
Abstract
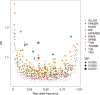
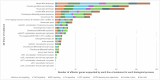
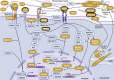
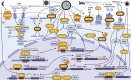
Osteoarthritis is the third most rapidly growing health condition associated with disability, after dementia and diabetes1. By 2050, the total number of patients with osteoarthritis is estimated to reach 1 billion worldwide2. As no disease-modifying treatments exist for osteoarthritis, a better understanding of disease aetiopathology is urgently needed. Here we perform a genome-wide association study meta-analyses across up to 489,975 cases and 1,472,094 controls, establishing 962 independent associations, 513 of which have not been previously reported. Using single-cell multiomics data, we identify signal enrichment in embryonic skeletal development pathways. We integrate orthogonal lines of evidence, including transcriptome, proteome and epigenome profiles of primary joint tissues, and implicate 700 effector genes. Within these, we find rare coding-variant burden associations with effect sizes that are consistently higher than common frequency variant associations. We highlight eight biological processes in which we find convergent involvement of multiple effector genes, including the circadian clock, glial-cell-related processes and pathways with an established role in osteoarthritis (TGFβ, FGF, WNT, BMP and retinoic acid signalling, and extracellular matrix organization). We find that 10% of the effector genes express a protein that is the target of approved drugs, offering repurposing opportunities, which can accelerate translation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: U.S., D.F.G., K. Stefansson, L. Stefánsdóttir, V.T. and G. Thorleifsson are employed by deCODE genetics/Amgen. M. Isijima has received research support from Stryker, Zimmer-biomet and Mathys; is a member of the editorial/governing board of the journal of joint surgery and research and osteoarthritis and cartilage; and is a board member and committee appointment for the osteoarthritis research society international. A. Baras, M.A.R.F., L. Lotta, M.J. and A.G.U. are employed at Regeneron Pharmaceuticals. A.M.V. is a consultant for Zoe Global. In the past 3 years, S.A.T. has received remuneration for scientific advisory board membership from Sanofi, GlaxoSmithKline, Foresite Labs and Qiagen. S.A.T. is a co-founder and holds equity in Transition Bio and Ensocell. From 8 January 2024, S.A.T. is a part-time employee of GlaxoSmithKline. O.N.F. has received fees for lecture by Heraeus Medical and Ortomedic AS the past three years. C.E. received unrestricted research grants from Novo Nordisk and Abbott Diagnostics; no personal fees. J.A. Singh has received consultant fees from ROMTech, Atheneum, Clearview healthcare partners, American College of Rheumatology, Yale, Hulio, Horizon Pharmaceuticals/DINORA, Frictionless Solutions, Schipher, Crealta/Horizon, Medisys, Fidia, PK Med, Two labs, Adept Field Solutions, Clinical Care options, Putnam associates, Focus forward, Navigant consulting, Spherix, MedIQ, Jupiter Life Science, UBM, Trio Health, Medscape, WebMD and Practice Point communications; the National Institutes of Health; and the American College of Rheumatology. J.A. Singh has received institutional research support from Zimmer Biomet Holdings. J.A. Singh received food and beverage payments from Intuitive Surgical/Philips Electronics North America. J.A. Singh owns stock options in Atai life sciences, Kintara therapeutics, Intelligent Biosolutions, Acumen pharmaceutical, TPT Global Tech, Vaxart pharmaceuticals, Atyu biopharma, Adaptimmune Therapeutics, GeoVax Labs, Pieris Pharmaceuticals, Enzolytics, Seres Therapeutics, Tonix Pharmaceuticals Holding, Aebona Pharmaceuticals and Charlotte’s Web Holdings. J.A. Singh previously owned stock options in Amarin, Viking and Moderna pharmaceuticals. J.A. Singh is on the speaker’s bureau of Simply Speaking. J.A. Singh was a member of the executive of Outcomes Measures in Rheumatology (OMERACT), an organization that develops outcome measures in rheumatology and receives arms-length funding from eight companies. J.A. Singh serves on the FDA Arthritis Advisory Committee. J.A. Singh is the co-chair of the Veterans Affairs Rheumatology Field Advisory Board (FAB). J.A. Singh is the editor and the Director of the University of Alabama at Birmingham (UAB) Cochrane Musculoskeletal Group Satellite Center on Network Meta-analysis. J.A. Singh previously served as a member of the following committees: the American College of Rheumatology’s (ACR) Annual Meeting Planning Committee (AMPC) and Quality of Care Committees, the Chair of the ACR Meet-the-Professor, Workshop and Study Group Subcommittee and the co-chair of the ACR Criteria and Response Criteria subcommittee. N.P.H. has received institutional support, lecturer’s fees or honoraria from Waldemar Link, Zimmer Biomet, DePuy Synthes and Heraeus Medical, and has a license agreement with Waldemar Link. The other authors declare no competing interests.
Figures




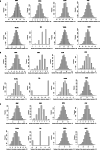
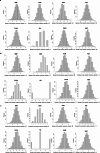
References
-
- GBD 2021 Diseases and Injuries Collaborators. Global incidence, prevalence, years lived with disability (YLDs), disability-adjusted life-years (DALYs), and healthy life expectancy (HALE) for 371 diseases and injuries in 204 countries and territories and 811 subnational locations, 1990–2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet403, 2133–2161 (2024). - PMC - PubMed
-
- Castaño-Betancourt, M. C. et al. The contribution of hip geometry to the prediction of hip osteoarthritis. Osteoarthr. Cartil.21, 1530–1536 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

