Ketomimetic nutrients remodel the glycocalyx and trigger a metabolic defense in breast cancer cells
- PMID: 40205476
- PMCID: PMC11984013
- DOI: 10.1186/s40170-025-00385-3
Ketomimetic nutrients remodel the glycocalyx and trigger a metabolic defense in breast cancer cells
Abstract
Background: While the triggers for the metastatic transformation of breast cancer (BC) cells remain unknown, recent evidence suggests that intrinsic cellular metabolism could be a crucial driver of migratory disposition and chemoresistance. Aiming to decipher the molecular mechanisms involved in BC cell metabolic maneuvering, we study how a ketomimetic (ketone body-rich, low glucose) nutrient medium can engineer the glycocalyx and metabolic signature of BC cells, to further maneuver their response to therapy.
Methods: Doxorubicin (DOX) has been used as a model chemotherapeutic in this study. Bioorthogonal imaging was used to assess the degree of sialylation of the glycocalyx along with measurements of drug-induced cytotoxicity and drug internalization. Single cell label-free metabolic imaging has been performed, coupled with measurement of cellular proliferative and migratory abilities, and MS-based metabolomic screens. Transcriptomic analysis of crucial enzymes was performed using total RNA extraction and rt-qPCR.
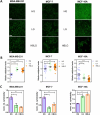
Results: We found an inverse correlation of glycocalyx sialylation with drug-induced cytotoxicity and drug internalization, where ketomimetic media enhanced sialylation and protected BC cells from DOX. These hypersialylated cells proliferated slower and migrated faster as compared to their counterparts receiving a high glucose media, while exhibiting a preference for glycolysis. These cells also showed pronounced lipid droplet accumulation coupled with an inversion in their metabolomic profile. Enzymatic removal of sialic acid moieties at the glycocalyx revealed for the first time, a direct role of sialic acids as defense guards, blocking DOX entry at the cellular membrane to curtail internalization. Interestingly, the non-cancerous mammary epithelial cells exhibited opposite trends and this differential pattern in cancer vs. normal cells was traced to its biochemical roots, i.e. the expression levels of key enzymes involved in sialylation and fatty acid synthesis.
Conclusions: Our findings revealed that a ketomimetic medium enhances chemoresistance and invasive disposition of BC cells via two main oncogenic pathways: hypersialylation and lipid synthesis. We propose that the crosstalk between these pathways, juxtaposed at the synthesis of the glycan precursor UDP-GlcNAc, furthers advancement of a metastatic phenotype in BC cells under ketomimetic conditions. Non-cancerous cells lack this dual defense machinery and end up being sensitized to DOX under ketomimetic conditions.
Keywords: Breast cancer metabolism; Glycocalyx engineering; Hypersialylation; Ketogenic diet; Ketomimetic; Lipid metabolism.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Update of
-
Ketomimetic Nutrients Trigger a Dual Metabolic Defense in Breast Cancer Cells.bioRxiv [Preprint]. 2024 Jul 31:2024.07.03.601966. doi: 10.1101/2024.07.03.601966. bioRxiv. 2024. Update in: Cancer Metab. 2025 Apr 9;13(1):18. doi: 10.1186/s40170-025-00385-3. PMID: 39005423 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

