Genomic and functional analysis of rmp locus variants in Klebsiella pneumoniae
- PMID: 40205597
- PMCID: PMC11984045
- DOI: 10.1186/s13073-025-01461-5
Genomic and functional analysis of rmp locus variants in Klebsiella pneumoniae
Abstract
Background: Klebsiella pneumoniae is an opportunistic pathogen and a leading cause of healthcare-associated infections in hospitals, which are frequently antimicrobial resistant (AMR). Exacerbating the public health threat posed by K. pneumoniae, some strains also harbour additional hypervirulence determinants typically acquired via mobile genetic elements such as the well-characterised large virulence plasmid KpVP-1. The rmpADC locus is considered a key virulence feature of K. pneumoniae and is associated with upregulated capsule expression and the hypermucoid phenotype, which can enhance virulence by contributing to serum resistance. Typically such strains have been susceptible to all antimicrobials besides ampicillin; however, the recent emergence of AMR hypermucoid strains is concerning.
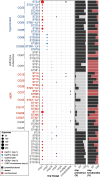
Methods: Here, we investigate the genetic diversity, evolution, mobilisation and prevalence of rmpADC, in a dataset of 14,000 genomes from isolates of the Klebsiella pneumoniae species complex, and describe the RmST virulence typing scheme for tracking rmpADC variants for the purposes of genomic surveillance. Additionally, we examine the functionality of representatives for variants of rmpADC introduced into a mutant strain lacking its native rmpADC locus.
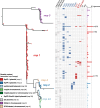
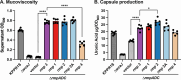
Results: The rmpADC locus was detected in 7% of the dataset, mostly from genomes of K. pneumoniae and a very small number of K. variicola and K. quasipneumoniae. Sequence variants of rmpADC grouped into five distinct lineages (rmp1, rmp2, rmp2A, rmp3 and rmp4) that corresponded to unique mobile elements, and were differentially distributed across different populations (i.e. clonal groups) of K. pneumoniae. All variants were demonstrated to produce enhanced capsule production and hypermucoviscosity.
Conclusions: These results provide an overview of the diversity and evolution of a prominent K. pneumoniae virulence factor and support the idea that screening for rmpADC in K. pneumoniae isolates and genomes is valuable to monitor the emergence and spread of hypermucoid K. pneumoniae, including AMR strains.
Keywords: Klebsiella pneumoniae; Genomic surveillance; Hypermucoid; Hypermucoviscosity; Hypervirulence; Virulence; Virulence plasmids.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

