Modeling a mesenchymal cell state by bioprinting for the molecular analysis of dormancy in melanoma
- PMID: 40206148
- PMCID: PMC11979991
- DOI: 10.1016/j.mtbio.2025.101674
Modeling a mesenchymal cell state by bioprinting for the molecular analysis of dormancy in melanoma
Abstract
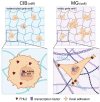
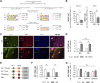
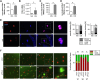
Malignant melanoma is a highly aggressive tumor originating from the pigment producing cells, the melanocytes. It accounts for the majority of skin cancer related deaths worldwide. This is often due to the development of therapy resistance or tumor dormancy, eventually resulting in tumor relapse by yet undefined mechanisms. Tumor dormancy is thought to be mediated by the cellular microenvironment and models taking this factor into account are urgently needed. We 3D bioprinted melanoma cells in the hydrogels Cellink Bioink (CIB) or Matrigel (MG), each as a substitute of the extracellular matrix, and, thereby, induced a quiescent or a proliferative phenotype of the melanoma cell lines, respectively. RNA-Seq with subsequent comprehensive bioinformatical and molecular analyses assigned CIB-cultured cells to a predominantly mesenchymal and Matrigel-cultured cells to a more mitotic phenotype, emphasizing the CIB model as a suitable platform for the investigation of dormancy under consideration of the microenvironment. Melanoma cells in CIB 3D culture reflect a quiescent and migratory active cell state e.g. by revealing significant downregulation of genes associated with replication and cell cycle progression in this setting. Using this model system, we identified the mechanosensory gene FHL2 as one early sensor of changes in the ECM and suggest a FHL2-p21/AP-1 axis contributing to the dormant phenotype of melanoma cells in CIB.
Keywords: 3D cell culture; Bioinformatics; Cancer; Mechanosensation; Quiescence.
© 2025 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Luzzi K.J., MacDonald I.C., Schmidt E.E., Kerkvliet N., Morris V.L., Chambers A.F., Groom A.C. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. - DOI - PMC - PubMed
-
- Schmid R., Schmidt S.K., Hazur J., Detsch R., Maurer E., Boccaccini A.R., Hauptstein J., Teßmar J., Blunk T., Schrüfer S., Schubert D.W., Horch R.E., Bosserhoff A.K., Arkudas A., Kengelbach-Weigand A. Comparison of hydrogels for the development of well-defined 3D cancer models of breast cancer and melanoma. Cancers. 2020;12:2320. doi: 10.3390/CANCERS12082320. 12 (2020) 2320. - DOI - PMC - PubMed
-
- Schmid R., Schmidt S.K., Detsch R., Horder H., Blunk T., Schrüfer S., Schubert D.W., Fischer L., Thievessen I., Heltmann-Meyer S., Steiner D., Schneidereit D., Friedrich O., Grüneboom A., Amouei H., Wajant H., Horch R.E., Bosserhoff A.K., Arkudas A., Kengelbach-Weigand A. A new printable alginate/hyaluronic acid/gelatin hydrogel suitable for biofabrication of in vitro and in vivo metastatic melanoma models. Adv. Funct. Mater. 2022;32 doi: 10.1002/ADFM.202107993. - DOI
LinkOut - more resources
Full Text Sources

