Structure, function and evolution of the bacterial DinG-like proteins
- PMID: 40206346
- PMCID: PMC11981726
- DOI: 10.1016/j.csbj.2025.03.023
Structure, function and evolution of the bacterial DinG-like proteins
Abstract
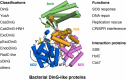
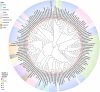
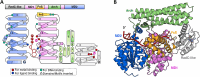
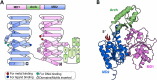
The damage-inducible G (DinG)-like proteins represent a widespread superfamily 2 (SF2) of DNA helicases, exhibiting remarkable diversity in domain architecture, substrate specificity, regulatory mechanisms, biological functions, interaction partners, and taxonomic distribution. Many characterized DinG-like proteins play critical roles in bacterial stress responses and immunity, including the SOS response, DNA repair, and phage interference. This review aims to provide a summary of bacterial DinG-like proteins, categorizing them into subgroups such as DinG, YoaA, CasDinG, CasDinG-HNH, ExoDinG, pExoDinG, EndoDinG, RadC-like DinG, sDinG, and others. This classification provides an analysis of sequence-structure-function relationships within this superfamily. Further sequence clustering revealed inter-cluster relationships and subgroup heterogeneity, suggesting potential functional divergence. Integrating sequence analysis, domain architecture, structural data, and genomic context enabled functional predictions for these DinG-like protein subgroups, shedding light on their evolutionary and biological significance.
Keywords: CRISPR interference; DNA repair; DinG; SOS response; Structure.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







References
-
- White M.F. Structure, function and evolution of the XPD family of iron-sulfur-containing 5′-> 3′ DNA helicases. Biochem Soc Trans. 2009;37(Pt 3):547–551. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
