Clinical and molecular overlap between nucleotide excision repair (NER) disorders and DYRK1A haploinsufficiency syndrome
- PMID: 40206408
- PMCID: PMC11979163
- DOI: 10.3389/fnins.2025.1554093
Clinical and molecular overlap between nucleotide excision repair (NER) disorders and DYRK1A haploinsufficiency syndrome
Abstract
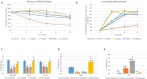
Nucleotide excision repair (NER) disorders are genetic conditions caused by defects in the pathway responsible for repairing DNA lesions due to UV radiation. These defects lead to a variety of heterogeneous disorders, including Cockayne syndrome (CS) and trichothiodystrophy (TTD). In this study, we report 11 patients initially suspected of having CS or TTD who were ultimately diagnosed with DYRK1A haploinsufficiency syndrome using high-throughput sequencing. Comparing clinical presentations, we observed that DYRK1A symptoms overlapped with CS, with shared features such as intellectual disability and microcephaly, systematically present in both disorders and other common symptoms including feeding difficulties, abnormal brain imaging, ataxic gait, hypertonia, and deep-set eyes. However, distinctive features of DYRK1A syndrome, such as severely impaired language, febrile seizures, and autistic behavior or anxiety, helped differentiate it from CS, which typically manifests with severe growth delay, bilateral cataracts, and pigmentary retinopathy. Among the cohort, three patients carried novel DYRK1A variants, including two truncating and one in-frame variant p.Val237_Leu241delinsGlu whose pathogenicity have been confirmed through functional analysis of DYRK1A protein. While previous research has implicated DYRK1A in DNA repair, with DYRK1A being one of the most downregulated genes in CS cells, our study found that DYRK1A patient-derived cell lines did not exhibit NER defects and did not share the CS transcriptomic signature. These findings suggest that if clinical symptoms overlap stems from common molecular disruptions, DYRK1A is involved downstream of the CS genes. This research highlights the importance of considering DYRK1A haploinsufficiency syndrome in the differential diagnoses for NER disorders.
Keywords: Cockayne syndrome; DYRK1A gene; ERCC6/CSB gene; ERCC8/CSA gene; nucleotide excision repair (NER); trichothiodystrophy.
Copyright © 2025 Le May, Courraud, Boujelbène, Obringer, Ogi, Lehmann, Laffargue, Lehalle, Mizuno, Mohammed, Ormières, Willems, Laugel and Calmels.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Bélanger F., Roussel C., Sawchyn C., St-Hilaire E., Gezzar-Dandashi S., Kimenyi Ishimwe A. B., et al. . (2023). A genome-wide screen reveals that Dyrk1A kinase promotes nucleotide excision repair by preventing aberrant overexpression of cyclin D1 and p21. J. Biol. Chem. 299:104900. doi: 10.1016/j.jbc.2023.104900, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

