Highly selective DNA aptamer sensor for intracellular detection of coenzyme A
- PMID: 40206557
- PMCID: PMC11976443
- DOI: 10.1039/d5sc00332f
Highly selective DNA aptamer sensor for intracellular detection of coenzyme A
Abstract
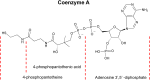
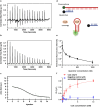
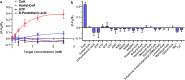
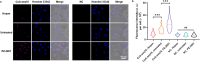
Detecting Coenzyme A (CoA) in cells is vital for understanding its role in metabolism. DNA aptamers, though widely used for monitoring many other molecules, have not been effective for CoA detection, as previous attempts at obtaining DNA aptamers for CoA using SELEX resulted in aptamers that only recognize the adenine moiety of CoA. This "tyranny" of adenine dominating in SELEX has, therefore, hampered the SELEX of aptamers specific for CoA. To meet this challenge, we employed a capture SELEX method by incorporating rigorous counter selections against adenine, adenosine, ATP, pantetheine, and pantothenic acid, resulting in a highly specific DNA aptamer for CoA over adenosine, ATP and other related metabolites such as NADH, with a dissociation constant of 48.9 μM. This aptamer was then converted to a fluorescent sensor for CoA across pH 6.4-8.0. Confocal microscopy showed its ability to visualize CoA in living cells, with fluorescence changes observed upon manipulating CoA levels. This method broadens SELEX's application and presents a promising approach for studying and understanding CoA dynamics.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

References
-
- Huizenga D. E. Szostak J. W. Biochemistry. 1995;34:656–665. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

