Eurasian 1C swine influenza A virus exhibits high pandemic risk traits
- PMID: 40207467
- PMCID: PMC12064114
- DOI: 10.1080/22221751.2025.2492210
Eurasian 1C swine influenza A virus exhibits high pandemic risk traits
Abstract
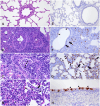
ABSTRACTRecent surveillance has identified an expansion of swine H1 1C influenza viruses in Eurasian swine. Since 2010, at least 21 spillover events of 1C virus into humans have been detected and three of these occurred from July to December of 2023. Pandemic risk assessment of H1 1C influenza virus revealed that individuals born after 1950 had limited cross-reactive antibodies, confirming that they are antigenically novel viruses. The 1C virus exhibited phenotypic signatures similar to the 2009 pandemic H1N1 virus, including human receptor preference, productive replication in human airway cells, and robust environmental stability. Efficient inter- and intraspecies airborne transmission using the swine and ferret models was observed, including efficient airborne transmission to ferrets with pre-existing human seasonal H1N1 immunity. Together our data suggest H1 1C influenza virus poses a relatively high pandemic risk.
Keywords: Influenza virus; pandemic risk assessment; pre-existing immunity; swine; transmission.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
