The PIN1-p38-CtIP signalling axis protects stalled replication forks from deleterious degradation
- PMID: 40207632
- PMCID: PMC11983131
- DOI: 10.1093/nar/gkaf278
The PIN1-p38-CtIP signalling axis protects stalled replication forks from deleterious degradation
Abstract
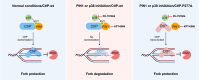
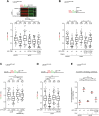
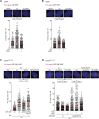
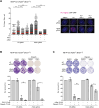
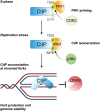
Human CtIP plays a critical role in homologous recombination (HR) by promoting the resection of DNA double-strand breaks. Moreover, CtIP maintains genome stability through protecting stalled replication forks from nucleolytic degradation. However, the upstream signalling mechanisms governing the molecular switch between these two CtIP-dependent processes remain largely elusive. Here, we show that phosphorylation of CtIP by the p38α stress kinase and subsequent PIN1-mediated CtIP cis-to-trans isomerization is required for fork stabilization but dispensable for HR. We found that stalled forks are degraded in cells expressing non-phosphorylatable CtIP or lacking PIN1-p38α activity, while expression of a CtIP trans-locked mutant overcomes the requirement for PIN1-p38α in fork protection. We further reveal that Brca1-deficient mammary tumour cells that have acquired PARP inhibitor (PARPi) resistance regain chemosensitivity after PIN1 or p38α inhibition. Collectively, our findings identify the PIN1-p38-CtIP signalling pathway as a critical regulator of replication fork integrity.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

