Pseudomonas aeruginosa population dynamics in a vancomycin-induced murine model of gastrointestinal carriage
- PMID: 40207916
- PMCID: PMC12077156
- DOI: 10.1128/mbio.03136-24
Pseudomonas aeruginosa population dynamics in a vancomycin-induced murine model of gastrointestinal carriage
Abstract
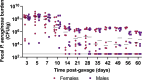
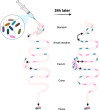
Pseudomonas aeruginosa is a common nosocomial pathogen and a major cause of morbidity and mortality in hospitalized patients. Multiple reports highlight that P. aeruginosa gastrointestinal colonization may precede systemic infections by this pathogen. Gaining a deeper insight into the dynamics of P. aeruginosa gastrointestinal carriage is an essential step in managing gastrointestinal colonization and could contribute to preventing bacterial transmission and progression to systemic infection. Here, we present a clinically relevant mouse model relying on parenteral vancomycin pretreatment and a single orogastric gavage of a controlled dose of P. aeruginosa. Robust carriage was observed with multiple clinical isolates, and carriage persisted for up to 60 days. Histological and microbiological examination of mice indicated that this model indeed represented carriage and not infection. We then used a barcoded P. aeruginosa library along with the sequence tag-based analysis of microbial populations (STAMPR) analytic pipeline to quantify bacterial population dynamics and bottlenecks during the establishment of the gastrointestinal carriage. Analysis indicated that most of the P. aeruginosa population was rapidly eliminated in the stomach, but the few bacteria that moved to the small intestine and the cecum expanded rapidly. Hence, the stomach constitutes a significant barrier against gastrointestinal carriage of P. aeruginosa, which may have clinical implications for hospitalized patients.
Importance: While Pseudomonas aeruginosa is rarely part of the normal human microbiome, carriage of the bacterium is quite frequent in hospitalized patients and residents of long-term care facilities. P. aeruginosa carriage is a precursor to infection. Options for treating infections caused by difficult-to-treat P. aeruginosa strains are dwindling, underscoring the urgency to better understand and impede pre-infection stages, such as colonization. Here, we use vancomycin-treated mice to model antibiotic-treated patients who become colonized with P. aeruginosa in their gastrointestinal tracts. We identify the stomach as a major barrier to the establishment of gastrointestinal carriage. These findings suggest that efforts to prevent gastrointestinal colonization should focus not only on judicious use of antibiotics but also on investigation into how the stomach eliminates orally ingested P. aeruginosa.
Keywords: Pseudomonas aeruginosa; STAMP; intestinal colonization; mouse; vancomycin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Update of
-
Pseudomonas aeruginosa population dynamics in a vancomycin-induced murine model of gastrointestinal carriage.bioRxiv [Preprint]. 2024 Aug 20:2024.08.19.608679. doi: 10.1101/2024.08.19.608679. bioRxiv. 2024. Update in: mBio. 2025 May 14;16(5):e0313624. doi: 10.1128/mbio.03136-24. PMID: 39229171 Free PMC article. Updated. Preprint.
Similar articles
-
Pseudomonas aeruginosa population dynamics in a vancomycin-induced murine model of gastrointestinal carriage.bioRxiv [Preprint]. 2024 Aug 20:2024.08.19.608679. doi: 10.1101/2024.08.19.608679. bioRxiv. 2024. Update in: mBio. 2025 May 14;16(5):e0313624. doi: 10.1128/mbio.03136-24. PMID: 39229171 Free PMC article. Updated. Preprint.
-
Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis.Cochrane Database Syst Rev. 2017 Apr 25;4(4):CD004197. doi: 10.1002/14651858.CD004197.pub5. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Jun 2;6:CD004197. doi: 10.1002/14651858.CD004197.pub6. PMID: 28440853 Free PMC article. Updated.
-
Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis.Cochrane Database Syst Rev. 2014 Nov 10;(11):CD004197. doi: 10.1002/14651858.CD004197.pub4. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2017 Apr 25;4:CD004197. doi: 10.1002/14651858.CD004197.pub5. PMID: 25383937 Updated.
-
Prophages are infrequently associated with antibiotic resistance in Pseudomonas aeruginosa clinical isolates.mSphere. 2025 Mar 25;10(3):e0090424. doi: 10.1128/msphere.00904-24. Epub 2025 Feb 13. mSphere. 2025. PMID: 39945525 Free PMC article.
-
Multi-omics profiling of cross-resistance between ceftazidime-avibactam and meropenem identifies common and strain-specific mechanisms in Pseudomonas aeruginosa clinical isolates.mBio. 2025 Jul 9;16(7):e0389624. doi: 10.1128/mbio.03896-24. Epub 2025 Jun 4. mBio. 2025. PMID: 40464559 Free PMC article.
References
-
- Ikuta KS, Swetschinski LR, Robles Aguilar G, Sharara F, Mestrovic T, Gray AP, Davis Weaver N, Wool EE, Han C, Gershberg Hayoon A, et al. . 2022. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet 400:2221–2248. doi:10.1016/S0140-6736(22)02185-7 - DOI - PMC - PubMed
-
- Centers for the Disease Control and Prevention . 2023. Pseudomonas aeruginosa infection. HAI | CDC. Available from: https://www.cdc.gov/hai/organisms/pseudomonas.html. Retrieved 8 March 2024.
-
- Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, et al. . 2014. Emerging infections program healthcare-associated infections and antimicrobial use prevalence survey team. N Engl J Med 370:1198–1208. doi:10.1056/NEJMoa1306801 - DOI - PMC - PubMed
-
- Centers for Disease Control and Prevention (U.S.) . 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention (U.S.)
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
