Whole Genome Sequencing and Genetic Diversity of Respiratory Viruses Detected in Children With Acute Respiratory Infections: A One-Year Cross-Sectional Study in Senegal
- PMID: 40207925
- PMCID: PMC11984337
- DOI: 10.1002/jmv.70342
Whole Genome Sequencing and Genetic Diversity of Respiratory Viruses Detected in Children With Acute Respiratory Infections: A One-Year Cross-Sectional Study in Senegal
Erratum in
-
Correction to "Whole Genome Sequencing and Genetic Diversity of Respiratory Viruses Detected in Children With Acute Respiratory Infections: A One-Year Cross-Sectional Study in Senegal".J Med Virol. 2025 May;97(5):e70419. doi: 10.1002/jmv.70419. J Med Virol. 2025. PMID: 40415274 Free PMC article. No abstract available.
Abstract
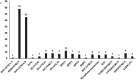
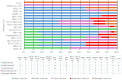
Acute respiratory infections (ARI) are a health priority, especially in countries with limited resources. They are a major cause of morbidity and mortality, especially among children and the elderly. In Senegal, the endemic circulation of respiratory viruses other than influenza has been demonstrated. However, there is a paucity of data exploring the genetic diversity of these viruses based on whole-genome sequencing. In this study, we present data on the genetic diversity of respiratory viruses in children under 15 years old in Senegal, including an overview of the different pathogens detected. Between November 2022 and November 2023, we collected nasopharyngeal swabs from children seen in curative consultations for symptoms of acute respiratory infections. Of the 156 children included, 73.7% tested positive for at least one pathogen. The most frequently detected virus was rhinovirus (50.0%), followed by influenza B (41.6%) and human parainfluenza virus type 3 (7.6%). Combinations of rhinovirus/influenza B, human parainfluenza virus type 2/human parainfluenza virus type 4, and rhinovirus/influenza B/adenovirus were the most frequently identified. A statistically significant association was detected between some of the viruses detected. A high genetic diversity of respiratory viruses circulating in children was revealed. The strains were phylogenetically close to various strains circulating worldwide, suggesting a global circulation of respiratory viruses. Our study provides the first complete genome sequences of human parainfluenza viruses type 2, 3, 4 and human bocavirus from Senegal and thus contributes to the enrichment of international databases on sequences from Senegal and underlines the importance of sequencing in the dynamics of pathogen circulation.
Keywords: NGS; Senegal; acute respiratory infections; children; virus; whole genome.
© 2025 The Author(s). Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Global Health Estimates: Leading Causes of Death. https://www.who.int/data/gho/data/themes/mortality-and-global-health-est..., accessed 7 May 2024.
-
- Annuaire_statistique_2019.pdf. https://www.sante.gouv.sn/sites/default/files/Annuaire_statistique_2019.pdf, accessed 13 May 2024.
-
- Fall A., Dieng A., Fallou Wade S., et al., “Children Under Five Years of Age in Senegal: A Group Highly Exposed to Respiratory Viruses Infections,” Virology: Research and Reviews 1 (2017): 10.15761/VRR.1000121, http://www.oatext.com/children-under-five-years-of-age-in-senegal-a-grou....
MeSH terms
LinkOut - more resources
Full Text Sources

