Recent emergence of cephalosporin-resistant Salmonella Typhi in India due to the endemic clone acquiring IncFIB(K) plasmid encoding blaCTX-M-15 gene
- PMID: 40208005
- PMCID: PMC12054180
- DOI: 10.1128/spectrum.00875-24
Recent emergence of cephalosporin-resistant Salmonella Typhi in India due to the endemic clone acquiring IncFIB(K) plasmid encoding blaCTX-M-15 gene
Abstract
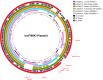
The emergence and spread of Salmonella Typhi (S. Typhi) resistant to third-generation cephalosporins is a serious global health concern. In this study, we genomically characterized 142 cephalosporin-resistant S. Typhi strains isolated from India. Comparative genome analysis revealed the emergence of a new clone of ceftriaxone-resistant S. Typhi harboring three plasmids of the incompatibility groups IncFIB(K), IncX1, and IncFIB(pHCM2). Among these, the IncFIB(K) plasmid confers resistance to third-generation cephalosporins through the blaCTX-M-15 gene, along with other resistance determinants such as aph(3"), aph(6'), sul2, dfrA14, qnrS, and tet(A). Phylogenetic analysis showed that the isolates from Gujarat (n = 140/142) belong to a distinct subclade (genotype 4.3.1.2.2) within genotype 4.3.1.2 (H58 lineage II). Single nucleotide polymorphism-based phylogenetic analysis of the core genes in IncFIB(K) suggested a close relatedness of the plasmid backbone to that of IncFIB(K) from other Enterobacteriales, indicating that H58 lineage II possesses the capability to acquire MDR plasmids from these organisms. This could indicate the potential onset of a new wave of ceftriaxone-resistant S. Typhi in India. The implementation of control measures-such as vaccination and improved water, sanitation, and hygiene systems-is crucial in areas where MDR or extensively drug-resistant S. Typhi strains are prevalent to curb the spread and impact of these resistant strains.
Importance: Typhoid fever remains a global health concern, especially in areas lacking sanitation and clean water. The rise of drug-resistant strains complicates treatment, increasing illness, death, and healthcare expenses. Travel facilitates the spread of these strains worldwide. Multidrug-resistant and extensively drug-resistant (XDR) strains, including those resistant to first-line antibiotics and fluoroquinolones, pose significant challenges. Azithromycin and third-generation cephalosporins are now preferred treatments. Recently, XDR typhoid emerged in Pakistan, resistant even to third-generation cephalosporins. India also faces challenges, with sporadic cases initially declining but now re-emerging. New strains in India show resistance to third-generation cephalosporins due to plasmid acquisition from other bacteria, particularly blaCTX-M-carrying IncFIB(K). Due to the ongoing nature of this outbreak, the data from this study deserve further consideration in order to control its spread in India.
Keywords: India; Salmonella Typhi; antimicrobial resistance; cephalosporin resistant; genome analysis; plasmid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- WHO . 2023. Typhoid. Available from: https://www.who.int/news-room/fact-sheets/detail/typhoid
-
- Mogasale V, Maskery B, Ochiai RL, Lee JS, Mogasale VV, Ramani E, Kim YE, Park JK, Wierzba TF. 2014. Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk-factor adjustment. Lancet Glob Health 2:e570–e580. doi:10.1016/S2214-109X(14)70301-8 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

