Species-specific components of the Helicobacter pylori Cag type IV secretion system
- PMID: 40208031
- PMCID: PMC12070742
- DOI: 10.1128/iai.00493-24
Species-specific components of the Helicobacter pylori Cag type IV secretion system
Abstract
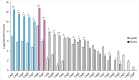
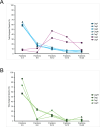
Helicobacter pylori strains containing the cag pathogenicity island (PAI) deliver an effector protein (CagA) and non-protein substrates into gastric cells through a process that requires the Cag type IV secretion system (T4SS). The Cag T4SS outer membrane core complex (OMCC) contains multiple copies of five proteins, two of which are species-specific proteins. By using modifications of a previously described OMCC immunopurification method and optimized mass spectrometric methods, we have now isolated additional cag PAI-encoded proteins that are present in lower relative abundance. Four of these proteins (CagW, CagL, CagI, and CagH) do not exhibit sequence relatedness to T4SS components in other bacterial species. Size exclusion chromatography analysis of immunopurified samples revealed that CagW, CagL, CagI, and CagH co-elute with OMCC components. These four Cag proteins are copurified with the OMCC in immunopurifications from a Δcag3 mutant strain (lacking peripheral OMCC components), but not from a ΔcagX mutant strain (defective in OMCC assembly). Negative stain electron microscopy analysis indicated that OMCC preparations isolated from ΔcagW, cagL::kan, ΔcagI, and ΔcagH mutant strains are indistinguishable from wild-type OMCCs. In summary, by using several complementary methods, we have identified multiple species-specific Cag proteins that are associated with the Cag T4SS OMCC and are required for T4SS activity.
Keywords: Helicobacter pylori; bacterial protein secretion; bacterial secretion systems; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. 2017. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153:420–429. doi:10.1053/j.gastro.2017.04.022 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

