A Salmonella enterica serovar Typhimurium genome-wide CRISPRi screen reveals a role for type 1 fimbriae in evasion of antibody-mediated agglutination
- PMID: 40208041
- PMCID: PMC12070745
- DOI: 10.1128/iai.00574-24
A Salmonella enterica serovar Typhimurium genome-wide CRISPRi screen reveals a role for type 1 fimbriae in evasion of antibody-mediated agglutination
Abstract
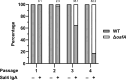
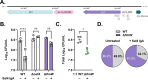
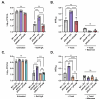
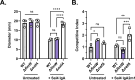
The O5-specific monoclonal IgA antibody, Sal4, mediates the conversion of Salmonella enterica serovar Typhimurium (STm) from virulent, free-swimming cells to non-motile, multicellular biofilm-like aggregates within a matter of hours. We hypothesize that the rapid transition from an invasive to a non-invasive state is an adaptation of STm to Sal4 IgA exposure. In this report, we performed a genome-wide CRISPR interference (CRISPRi) screen to identify STm genes that influence multicellular aggregate formation in response to Sal4 IgA treatment. From a customized library of >36,000 spacers, ~1% (373) were enriched at the top of the culture supernatant after two consecutive rounds of Sal4 IgA treatment. The enriched spacers mapped to a diversity of targets, including genes involved in O-antigen modification, cyclic-di-GMP metabolism, outer membrane biosynthesis/signaling, and invasion/virulence, with the most frequently targeted gene being fimW, which encodes a negative regulator of type 1 fimbriae (T1F) expression. Generation of a STm ΔfimW strain confirmed that the loss of FimW activity results in a hyperfimbriated phenotype and evasion of Sal4 IgA-mediated agglutination in solution. Closer examination of the fimW mutant revealed its propensity to form biofilms at the air-liquid interface in response to Sal4 exposure, suggesting that T1F "primes" STm to transition from a planktonic to a sessile state, possibly by facilitating bacterial attachment to abiotic surfaces. These findings shed light on the mechanism by which IgA antibodies influence STm virulence in the intestinal environment.
Keywords: Salmonella; adhesion; antibody; immunity; mucosal.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Distinct adaptation and epidemiological success of different genotypes within Salmonella enterica serovar Dublin.Elife. 2025 Jun 25;13:RP102253. doi: 10.7554/eLife.102253. Elife. 2025. PMID: 40560760 Free PMC article.
-
The sulfur assimilation pathway mitigates redox stress from acidic pH in Salmonella Typhi H58.mBio. 2025 Jul 9;16(7):e0046725. doi: 10.1128/mbio.00467-25. Epub 2025 May 27. mBio. 2025. PMID: 40422406 Free PMC article.
-
Functional analysis of cyclic diguanylate-modulating proteins in Vibrio fischeri.mSystems. 2024 Nov 19;9(11):e0095624. doi: 10.1128/msystems.00956-24. Epub 2024 Oct 22. mSystems. 2024. PMID: 39436151 Free PMC article.
-
Uncommon Non-MS Demyelinating Disorders of the Central Nervous System.Curr Neurol Neurosci Rep. 2025 Jul 1;25(1):45. doi: 10.1007/s11910-025-01432-8. Curr Neurol Neurosci Rep. 2025. PMID: 40591029 Review.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
References
-
- Troeger C, Forouzanfar M, Rao PC, Khalil I, Brown A, Reiner RC, Fullman N, Thompson RL, Abajobir A, Ahmed M, et al. . 2017. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the global burden of disease study 2015. Lancet Infect Dis 17:909–948. doi:10.1016/S1473-3099(17)30276-1 - DOI - PMC - PubMed
-
- Mather AE, Reid SWJ, Maskell DJ, Parkhill J, Fookes MC, Harris SR, Brown DJ, Coia JE, Mulvey MR, Gilmour MW, Petrovska L, de Pinna E, Kuroda M, Akiba M, Izumiya H, Connor TR, Suchard MA, Lemey P, Mellor DJ, Haydon DT, Thomson NR. 2013. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341:1514–1517. doi:10.1126/science.1240578 - DOI - PMC - PubMed
-
- Feasey NA, Masesa C, Jassi C, Faragher EB, Mallewa J, Mallewa M, MacLennan CA, Msefula C, Heyderman RS, Gordon MA. 2015. Three epidemics of invasive multidrug-resistant Salmonella bloodstream infection in Blantyre, Malawi, 1998-2014. Clin Infect Dis 61 Suppl 4:S363–71. doi:10.1093/cid/civ691 - DOI - PMC - PubMed
-
- Okoro CK, Barquist L, Connor TR, Harris SR, Clare S, Stevens MP, Arends MJ, Hale C, Kane L, Pickard DJ, Hill J, Harcourt K, Parkhill J, Dougan G, Kingsley RA. 2015. Signatures of adaptation in human invasive Salmonella Typhimurium ST313 populations from sub-Saharan Africa. PLoS Negl Trop Dis 9:e0003611. doi:10.1371/journal.pntd.0003611 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

