Profiling the nasopharyngeal Microbiome in patients with community-acquired pneumonia caused by Streptococcus pneumoniae: diagnostic challenges and ecological insights
- PMID: 40208342
- PMCID: PMC11985632
- DOI: 10.1007/s00430-025-00828-0
Profiling the nasopharyngeal Microbiome in patients with community-acquired pneumonia caused by Streptococcus pneumoniae: diagnostic challenges and ecological insights
Abstract
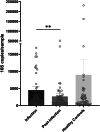
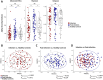
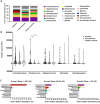
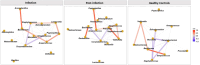
Community-acquired pneumonia (CAP) is a significant health threat for adults. Although conjugate vaccines have reduced pneumococcal CAP incidence in children, Streptococcus pneumoniae-related CAP remains prevalent among older adults. The nasopharynx acts as a reservoir for S. pneumoniae, yet the interplay between this pathogen and the nasopharyngeal microbiome during and after pneumonia remains poorly understood. This study included 61 adult patients diagnosed with pneumococcal CAP and 61 matched healthy controls. An S. pneumoniae-specific PCR, urine antigen tests and bacterial cultures were performed. Nasopharyngeal swabs collected at admission and three months post-infection were analyzed for microbiome dynamics through 16 S rRNA gene amplicon sequencing. 16 S rRNA gene amplicon sequencing revealed Streptococcus spp. in the majority of all nasopharyngeal samples during infection compared to the other diagnostic test performed. While overall bacterial biomass did not differ between groups, patients exhibited higher alpha diversity (p = 0.012) and lower microbiome stability post-infection. Beta diversity analysis distinguished infection from healthy status (p = 0.002). Taxonomic analysis showed similar core microbiota across groups, but Streptococcus spp. was significantly more abundant during infection, particularly in those patients with viral co-infections. Notably, unique significant bacterial interactions were identified both during and after infection, as well as in healthy states. A negative correlation was observed between Corynebacterium and Streptococcus spp. in infected patients, suggesting a potential antagonistic interaction between these taxa. The nasopharyngeal microbiome in patients with pneumococcal CAP demonstrates persistent disruption post-infection, characterized by lower resilience three months after acute illness. Additionally, we identified specific bacterial interplays during and after infection that differed from those in healthy donors. These bacterial dynamics might play critical roles in pathogen colonization resistance and infection prevention. Thus, our findings highlight the need for further investigation into microbial interactions and potential microbiome-based therapies for respiratory infections, particularly in vulnerable populations.
Keywords: Streptococcus pneumoniae; Community acquired pneumonia (CAP); Microbial interactions; Microbiome; Nasopharynx; Resilience; Viral infection.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
-
- Welte T, Torres A, Nathwani D (2012) Clinical and economic burden of community-acquired pneumonia among adults in Europe. Thorax 67(1):71–79 - PubMed
-
- McCullers JA (2014) The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 12(4):252–262 - PubMed
-
- Bogaert D, de Groot R, Hermans P (2004) Streptococcus pneumoniae colonisation: the key to Pneumococcal disease. Lancet Infect Dis 4(3):144–154 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

