Multi-channel smFRET study reveals a compact conformation of EF-G on the ribosome
- PMID: 40210088
- PMCID: PMC12175753
- DOI: 10.1016/j.biocel.2025.106782
Multi-channel smFRET study reveals a compact conformation of EF-G on the ribosome
Abstract
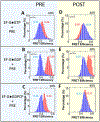
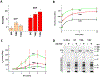
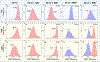
While elongation factor G (EF-G) is crucial for ribosome translocation, the role of its GTP hydrolysis remains ambiguous. EF-G's indispensability is further exemplified by the phosphorylation of human eukaryotic elongation factor 2 (eEF2) at Thr56, which inhibits protein synthesis globally, but its exact mechanism is not clear. In this study, we developed a multi-channel single-molecule FRET (smFRET) microscopy methodology to examine the conformational changes of E. coli EF-G induced by mutations that closely aligned with eEF2's Thr56 residue. We utilized Alexa 488/594 double-labeled EF-G to catalyze the translocation of fMet-Phe-tRNAPhe-Cy3 inside Cy5-L27 labeled ribosomes, allowing us to probe both processes within the same complex. Our findings indicate that in the presence of either GTP or GDPCP, wild-type EF-G undergoes a conformational extension upon binding to the ribosome to promote normal translocation. On the other hand, the T48E and T48V mutations did not affect GTP/GDP binding or GTP hydrolysis, but impeded Poly(Phe) synthesis and caused EF-G to adopt a unique compact conformation, which was not observed when the mutants interact solely with the SRL. This study provides new insights into EF-G's adaptability and sheds light on the modification mechanism of human eEF2.
Keywords: Compact EF-G; Induced-fit; Multi-channel smFRET; Ribosome translocation; Single molecule FRET.
Copyright © 2025 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Update of
-
Multi-Channel smFRET study reveals a Compact conformation of EF-G on the Ribosome.bioRxiv [Preprint]. 2024 Jan 28:2024.01.27.577133. doi: 10.1101/2024.01.27.577133. bioRxiv. 2024. Update in: Int J Biochem Cell Biol. 2025 Jul;184:106782. doi: 10.1016/j.biocel.2025.106782. PMID: 38328191 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

