Plasma-to-tumour tissue integrated proteomics using nano-omics for biomarker discovery in glioblastoma
- PMID: 40210624
- PMCID: PMC11986092
- DOI: 10.1038/s41467-025-58252-0
Plasma-to-tumour tissue integrated proteomics using nano-omics for biomarker discovery in glioblastoma
Abstract
Glioblastoma (GB) is the most lethal brain cancer, with patient survival rates remaining largely unchanged over the past two decades. Here, we introduce the Nano-omics integrative workflow that links systemic (plasma) and localised (tumour tissue) protein changes associated with GB progression. Mass spectrometry analysis of the nanoparticle biomolecule corona in GL261-bearing mice at different stages of GB revealed plasma protein alterations, even at low tumour burden, with over 30% overlap between GB-specific plasma and tumour tissue proteomes. Analysis of matched plasma and surgically resected tumour samples from high-grade glioma patients demonstrates the clinical applicability of the Nano-omics pipeline. Cross-species correlation identified 48 potential GB biomarker candidates involved in actin cytoskeleton organisation, focal adhesion, platelet activation, leukocyte migration, amino acid biosynthesis, carbon metabolism, and phagosome pathways. The Nano-omics approach holds promise for the discovery of early detection and disease monitoring biomarkers of central nervous system conditions, paving the way for subsequent clinical validation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
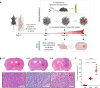
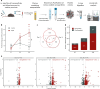
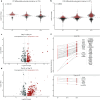
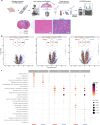
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

