Constraints on the optimization of gene product diversity
- PMID: 40210719
- PMCID: PMC12048591
- DOI: 10.1038/s44320-025-00095-4
Constraints on the optimization of gene product diversity
Abstract
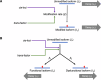
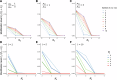
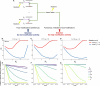
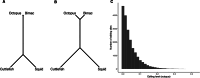
RNA and proteins can have diverse isoforms due to post-transcriptional and post-translational modifications. A fundamental question is whether these isoforms are mostly beneficial or the result of noisy molecular processes. To assess the plausibility of these explanations, we developed mathematical models depicting different regulatory architectures and investigated isoform evolution under multiple population genetic regimes. We found that factors beyond selection, such as effective population size and the number of cis-acting loci, significantly influence evolutionary outcomes. We found that sub-optimal phenotypes are more likely to evolve when populations are small and/or when the number of cis-loci is large. We also discovered that opposing selection on cis- and trans-acting loci can constrain adaptation, leading to a non-monotonic relationship between effective population size and optimization. More generally, our models provide a quantitative framework for developing statistical tests to analyze empirical data; as a demonstration of this, we analyzed A-to-I RNA editing levels in coleoids and found these to be largely consistent with non-adaptive explanations.
Keywords: Constraint; Evolutionary Theory; Gene Product Diversity; Optimization; Post-transcriptional Modification.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures
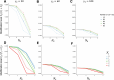
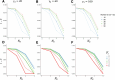

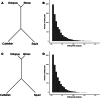
Update of
-
Genetic and selective constraints on the optimization of gene product diversity.bioRxiv [Preprint]. 2024 Jul 22:2024.07.17.603951. doi: 10.1101/2024.07.17.603951. bioRxiv. 2024. Update in: Mol Syst Biol. 2025 May;21(5):472-491. doi: 10.1038/s44320-025-00095-4. PMID: 39091777 Free PMC article. Updated. Preprint.
Similar articles
-
Genetic and selective constraints on the optimization of gene product diversity.bioRxiv [Preprint]. 2024 Jul 22:2024.07.17.603951. doi: 10.1101/2024.07.17.603951. bioRxiv. 2024. Update in: Mol Syst Biol. 2025 May;21(5):472-491. doi: 10.1038/s44320-025-00095-4. PMID: 39091777 Free PMC article. Updated. Preprint.
-
A mathematical framework for evo-devo dynamics.Theor Popul Biol. 2024 Feb;155:24-50. doi: 10.1016/j.tpb.2023.11.003. Epub 2023 Dec 2. Theor Popul Biol. 2024. PMID: 38043588
-
Diverse selective regimes shape genetic diversity at ADAR genes and at their coding targets.RNA Biol. 2015;12(2):149-61. doi: 10.1080/15476286.2015.1017215. RNA Biol. 2015. PMID: 25826567 Free PMC article.
-
The molecular population genetics of regulatory genes.Mol Ecol. 2000 Oct;9(10):1451-61. doi: 10.1046/j.1365-294x.2000.01016.x. Mol Ecol. 2000. PMID: 11050541 Review.
-
Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation.Mol Ecol. 2016 Jan;25(1):79-88. doi: 10.1111/mec.13288. Epub 2015 Jul 27. Mol Ecol. 2016. PMID: 26108992 Review.
Cited by
-
Alternative Mutational Architectures Producing Identical M-Matrices can Lead to Different Patterns of Evolutionary Divergence.Genome Biol Evol. 2025 May 30;17(6):evaf099. doi: 10.1093/gbe/evaf099. Genome Biol Evol. 2025. PMID: 40410123 Free PMC article.
References
-
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ (2012) The evolutionary landscape of alternative splicing in vertebrate species. Science 338(6114):1587–1593 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

