Single-cell profiling of peripheral blood mononuclear cells from patients treated with oncolytic adenovirus TILT-123 reveals baseline immune status as a predictor of therapy outcomes
- PMID: 40211048
- PMCID: PMC12183079
- DOI: 10.1038/s41417-025-00901-z
Single-cell profiling of peripheral blood mononuclear cells from patients treated with oncolytic adenovirus TILT-123 reveals baseline immune status as a predictor of therapy outcomes
Abstract
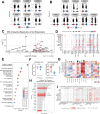
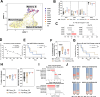
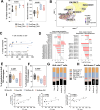
Oncolytic adenovirus Ad5/3-E2F-d24-hTNFa-IRES-hIL2 (TILT-123, igrelimogene litadenorepvec) shows promise as a therapeutic agent capable of causing tumor regression and activating host immunity. A phase I clinical study TUNIMO (NCT04695327) assessed its safety as monotherapy in patients with various solid tumors. Through single-cell profiling of peripheral blood, we identified distinct immunological features distinguishing responders from non-responders. Specifically, at baseline, responders demonstrated enhanced cytotoxic markers and stronger immune cell communication networks. Moreover, higher baseline CD16+ monocytes correlated with improved survival, while elevated regulatory T cells predicted poor response. T and B cell evaluation revealed contrasting patterns: responders showed higher numbers of T cells with predicted specificity to both adenovirus and tumor antigens, while elevated total memory B cells, regardless of specificity, predicted poor survival. Several T and B cell receptor segments matched those previously reported in other viral infections, suggesting possible cross-reactive immune responses. These findings emphasize that comprehensive biomarker analysis of peripheral blood should include not only cell frequencies but also transcriptional changes and distinct patterns of cellular and humoral immunity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: JHAC, CK, JMS and VC-C are employees and shareholders of TILT Biotherapeutics Ltd. DCAQ, LH, SS and RH are employees of TILT Biotherapeutics Ltd. AH is an employee and shareholder in TILT Biotherapeutics Ltd and shareholder in Circio Holdings ASA. All other authors declare no competing interests. Ethics approval and consent to participate: The TUNIMO trial protocol and ethics were reviewed by the Finnish Medical Agency and the Helsinki University Hospital Ethics board (approval 49/2020 and statement HUS/1804/2020). All patients gave written informed consent for participation. All study procedures were conducted in accordance with the relevant guidelines and regulations. Consent for publication: All patients gave written informed consent for publication of their clinical information.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

