Single-cell RNA sequencing reveals sex differences in the subcellular composition and associated gene-regulatory network activity of human carotid plaques
- PMID: 40211055
- PMCID: PMC11994450
- DOI: 10.1038/s44161-025-00628-y
Single-cell RNA sequencing reveals sex differences in the subcellular composition and associated gene-regulatory network activity of human carotid plaques
Abstract
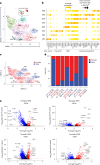
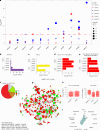
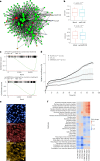
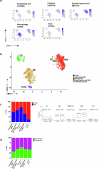
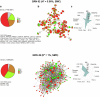
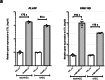
Carotid stenosis causes ischemic stroke in both sexes, but the clinical presentation and plaque characteristics differ. Here we run deep single-cell sequencing of 7,690 human carotid plaque cells from male and female patients. While we found no sex differences in major cell types, we identified a predominance of the osteogenic phenotype in smooth muscle cells, immunomodulating macrophages (MPs) and endothelial cells (ECs) undergoing endothelial-to-mesenchymal transition in females. In males, we found smooth muscle cells with the chondrocytic phenotype, MPs involved in tissue remodeling and ECs with angiogenic activity. Sex-biased subcellular clusters were integrated with tissue-specific gene-regulatory networks (GRNs) from the Stockholm-Tartu Atherosclerosis Reverse Network Engineering Task study. We identified GRN195 involved in angiogenesis and T cell-mediated cytotoxicity in male ECs, while in females, we found GRN33 and GRN122 related to TREM2-/TREM1+ MPs and endothelial-to-mesenchymal transition. The impact of GRN195 on EC proliferation in males was functionally validated, providing evidence for potential therapy targets for atherosclerosis that are sex specific.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.L.M.B. and A.R. are shareholders of Clinical Gene Networks AB (CGN) that has an invested interest in STARNET. C.L.M. has received funding from AstraZeneca for an unrelated project. All other authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
