The bridge-like lipid transport protein VPS13C/PARK23 mediates ER-lysosome contacts following lysosome damage
- PMID: 40211074
- PMCID: PMC12081312
- DOI: 10.1038/s41556-025-01653-6
The bridge-like lipid transport protein VPS13C/PARK23 mediates ER-lysosome contacts following lysosome damage
Abstract
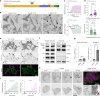
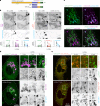
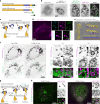
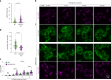
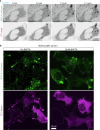
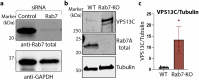
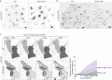
Based on genetic studies, lysosome dysfunction is thought to play a pathogenetic role in Parkinson's disease. Here we show that VPS13C, a bridge-like lipid-transport protein and a Parkinson's disease gene, is a sensor of lysosome stress or damage. Following lysosome membrane perturbation, VPS13C rapidly relocates from the cytosol to the surface of lysosomes where it tethers their membranes to the ER. This recruitment depends on Rab7 and requires a signal at the damaged lysosome surface that releases an inhibited state of VPS13C, which hinders access of its VAB domain to lysosome-bound Rab7. Although another Parkinson's disease protein, LRRK2, is also recruited to stressed or damaged lysosomes, its recruitment occurs at much later stages and by different mechanisms. Given the role of VPS13 proteins in bulk lipid transport, these findings suggest that lipid delivery to lysosomes by VPS13C is part of an early protective response to lysosome damage.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.D.C. is a member of the Scientific Advisory Board of Casma Therapeutics. The other authors declare no competing interests.
Figures










Update of
-
Lysosome damage triggers acute formation of ER to lysosomes membrane tethers mediated by the bridge-like lipid transport protein VPS13C.bioRxiv [Preprint]. 2025 Feb 26:2024.06.08.598070. doi: 10.1101/2024.06.08.598070. bioRxiv. 2025. Update in: Nat Cell Biol. 2025 May;27(5):776-789. doi: 10.1038/s41556-025-01653-6. PMID: 38895395 Free PMC article. Updated. Preprint.
References
-
- Schormair, B. et al. Diagnostic exome sequencing in early-onset Parkinson’s disease confirms VPS13C as a rare cause of autosomal-recessive Parkinson’s disease. Clin. Genet.93, 603–612 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- PF-RCE-1946/Parkinson's Foundation (Parkinson's Foundation, Inc.)
- NS36251/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- MC_UU_00018/1/RCUK | Medical Research Council (MRC)
- P30 DA018343/DA/NIDA NIH HHS/United States
- R01 NS036251/NS/NINDS NIH HHS/United States
- R01 GM105718/GM/NIGMS NIH HHS/United States
- GM105718/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- R37 NS036251/NS/NINDS NIH HHS/United States
- DA018343/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
LinkOut - more resources
Full Text Sources
Research Materials

