Identification of phloem-specific proteinaSEOus structure heterogeneity in sieve element of Populus trichocarpa
- PMID: 40211162
- PMCID: PMC11983751
- DOI: 10.1186/s12870-025-06439-4
Identification of phloem-specific proteinaSEOus structure heterogeneity in sieve element of Populus trichocarpa
Abstract
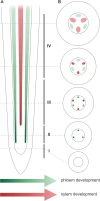
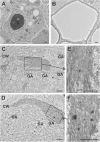
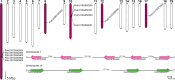
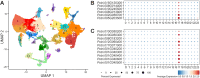
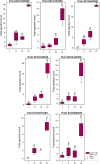
Phloem, an exceptional plant vascular tissue, facilitates the transport of photoassimilates, RNAs, and other signaling substances from the leaves to the roots throughout the plant. Among the specialized phloem cells are the conductive sieve elements (SEs), which are unique in that they remain alive despite lacking several cell organelles, including the nucleus, plastids, and most mitochondria. These SEs contain a specific proteinaceous structure composed of phloem-specific proteins (P-proteins), whose function is not yet fully understood. Various P-proteins have been characterized in broad range of model species, including Arabidopsis thaliana, and reported in Fabaceae and Cucurbitaceae plants. To date, only one P-protein has been identified in the model tree species Populus trichocarpa. Given the presence of multiple P-protein encoding genes across numerous plant species, we hypothesized the existence of multiple such genes in the Populus genome. Our genomic analysis uncovered 12 genes being potential orthologues to one of A. thaliana P-protein - SEOR (sieve element occlusion-related) genes, which may contribute to the proteinaceous structures observed in differentiating sieve elements. Our transcriptomic and proteomic analyses confirmed the expression of at least seven of these genes, indicating that the protein structure visible in mature sieve elements in P. trichocarpa may be heterogeneous.
Keywords: Populus trichocarpa; P-protein; Phloem; SEO; SEOR; Sieve element.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

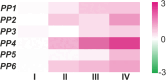
References
-
- Evert RF. Esau’s Plant Anatomy: John Wiley & Sons. New Jersey: Ltd; 2006.
-
- Aikman D. Phloem transport. Nature. 1974;252:760. - DOI
-
- Behnke HD, Sjolund RD. Sieve Elements: comparative structure, introduction and development. Berlin: Springer; 1990.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

