Unveiling polyphenol-protein interactions: a comprehensive computational analysis
- PMID: 40211304
- PMCID: PMC11983793
- DOI: 10.1186/s13321-025-00997-3
Unveiling polyphenol-protein interactions: a comprehensive computational analysis
Abstract
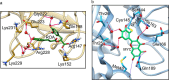
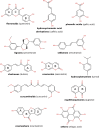
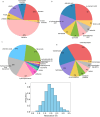
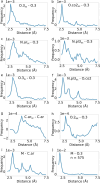
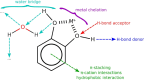
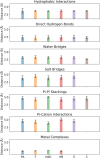
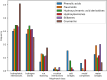
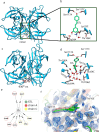
Our study investigates polyphenol-protein interactions, analyzing their structural diversity and dynamic behavior. Analysis of the entire Protein Data Bank reveals diverse polyphenolic structures, engaging in various noncovalent interactions with proteins. Interactions observed across crystal structures among diverse polyphenolic classes reveal similarities, underscoring consistent patterns across a spectrum of structural motifs. On the other hand, molecular dynamics (MD) simulations of polyphenol-protein complexes unveil dynamic binding patterns, highlighting the influx of water molecules into the binding site and underscoring limitations of static crystal structures. Water-mediated interactions emerge as crucial in polyphenol-protein binding, leading to variable binding patterns observed in MD simulations. Comparison of high- and low-resolution crystal structures as starting points for MD simulations demonstrates their robustness, exhibiting consistent dynamics regardless of the quality of the initial structural data. Additionally, the impact of glycosylation on polyphenol binding is explored, revealing its role in modulating interactions with proteins. In contrast to synthetic drugs, polyphenol binding seems to exhibit heightened flexibility, driven by dynamic water-mediated interactions, which may also facilitate their promiscuous binding. Comprehensive dynamic studies are, therefore essential to understand polyphenol-protein recognition mechanisms. Overall, our study provides novel insights into polyphenol-protein interactions, informing future research for harnessing polyphenolic therapeutic potential through rational drug design.Scientific contribution: In this study, we present an analysis of (natural) polyphenol-protein binding conformations, leveraging the entirety of the Protein Data Bank structural data on polyphenols, while extending the binding conformation sampling through molecular dynamics simulations. For the first time, we introduce experimentally supported large-scale systematization of polyphenol binding patterns. Moreover, our insight into the significance of explicit water molecules and hydrogen-bond bridging rationalizes the polyphenol promiscuity paradigm, advocating for a deeper understanding of polyphenol recognition mechanisms crucial for informed natural compound-based drug design.
Keywords: Dynamic behavior; Glycosylation; Molecular dynamics simulations; Noncovalent interactions; Polyphenol-protein complexes; Polyphenols; Water-mediated interactions.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Molecular dynamics and quantum mechanics of RNA: conformational and chemical change we can believe in.Acc Chem Res. 2010 Jan 19;43(1):40-7. doi: 10.1021/ar900093g. Acc Chem Res. 2010. PMID: 19754142 Free PMC article.
-
Exploiting the potential of natural polyphenols as antivirals against monkeypox envelope protein F13 using machine learning and all-atoms MD simulations.Comput Biol Med. 2023 Aug;162:107116. doi: 10.1016/j.compbiomed.2023.107116. Epub 2023 Jun 4. Comput Biol Med. 2023. PMID: 37302336 Free PMC article.
-
Prediction of Ordered Water Molecules in Protein Binding Sites from Molecular Dynamics Simulations: The Impact of Ligand Binding on Hydration Networks.J Chem Inf Model. 2018 Feb 26;58(2):350-361. doi: 10.1021/acs.jcim.7b00520. Epub 2018 Feb 5. J Chem Inf Model. 2018. PMID: 29308882 Free PMC article.
-
Analytical techniques for the study of polyphenol-protein interactions.Crit Rev Food Sci Nutr. 2017 Jul 3;57(10):2144-2161. doi: 10.1080/10408398.2015.1052040. Crit Rev Food Sci Nutr. 2017. PMID: 26566184 Review.
-
Molecular dynamics simulations to understand glycosaminoglycan interactions in the free- and protein-bound states.Curr Opin Struct Biol. 2022 Jun;74:102356. doi: 10.1016/j.sbi.2022.102356. Epub 2022 Mar 17. Curr Opin Struct Biol. 2022. PMID: 35306321 Free PMC article. Review.
Cited by
-
Modeling the Anti-Adhesive Role of Punicalagin Against Listeria Monocytogenes from the Analysis of the Interaction Between Internalin A and E-Cadherin.Int J Mol Sci. 2025 Jul 29;26(15):7327. doi: 10.3390/ijms26157327. Int J Mol Sci. 2025. PMID: 40806459 Free PMC article.
References
-
- Tijjani H, Zangoma MH, Mohammed ZS, Obidola SM, Egbuna C, Abdulai SI (2020) Polyphenols: classifications, biosynthesis and bioactivities. In: Egbuna C, Dable Tupas G (eds) Functional foods and nutraceuticals: bioactive components, formulations and innovations. Springer International Publishing, Cham, pp 389–414
-
- Belščak-Cvitanovic A, Durgo K, Hudek A, Bačun-Družina V, Komes D (2018) 1 - overview of polyphenols and their properties. In: Galanakis CM (ed) Polyphenols: properties, recovery, and applications. Woodhead Publishing, Sawston, pp 3–44
-
- Rana A, Samtiya M, Dhewa T, Mishra V, Aluko RE (2022) Health benefits of polyphenols: a concise review. J Food Biochem 46(10):e14264. 10.1111/jfbc.14264 - PubMed
-
- Quideau S, Deffieux D, Douat-Casassus C, Pouységu L (2011) Plant polyphenols: chemical properties, biological activities, and synthesis. Angewandte Chemie Int Ed 50(3):586–621. 10.1002/anie.201000044 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

