Acetylation suppresses breast cancer progression by sustaining CLYBL stability
- PMID: 40211376
- PMCID: PMC11984010
- DOI: 10.1186/s12967-025-06200-3
Acetylation suppresses breast cancer progression by sustaining CLYBL stability
Abstract
Background: The incidence of breast cancer remains high and it remains the leading cause of cancer-related deaths in women. A better understanding of the molecular mechanisms of breast cancer and identifying novel biomarkers will help improve therapeutic strategies. Citrate lyase beta like (CLYBL) is expressed at low levels in breast cancer tissues and is associated with low patient survival rates. In this study, we explored the regulatory mechanisms of CLYBL and its acetylation in breast cancer.
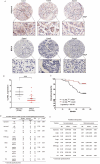
Methods: CLYBL expression patterns in breast cancer were assessed using a breast cancer tissue microarray, immunohistochemistry, and publicly available datasets. The acetylation patterns of CLYBL and the related regulatory functions were detected by high resolution mass spectrometry, immunoprecipitation assays, and western blot analysis. The potential effects of CLYBL and its acetylation on breast cancer were determined using both in vitro and in vivo assays.
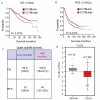
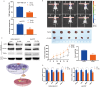
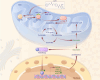
Results: CLYBL was expressed at lower levels in breast cancer samples compared with normal tissues. This low CLYBL expression was associated with poor patient survival rates. Overexpressing CLYBL could inhibit breast cancer and reduce NRF2 pathway-mediated antioxidants. We identified two acetylated lysine sites in CLYBL, K57 and K82, using acetylated peptide affinity enrichment and high-resolution mass spectrometry. Our results suggest that K82 is the main acetylation site. Further work showed that the p300/CBP associated factor (PCAF) and histone deacetylase 3 (HDAC3) as the CLYBL acetyltransferase and deacetylase, respectively. Additionally, CLYBL acetylation facilitates its own protein stability by reducing it affinity for ubiquitin, thus enhancing the anti-breast cancer effects.
Conclusion: We revealed the role of CLYBL overexpression and its acetylation in breast cancer. Our study suggests that CLYBL is a potential molecular target for breast cancer therapy.
Keywords: Acetylation; Breast cancer (BC); CLYBL; HDAC3; PCAF.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The experiments were performed according to the approved guidelines established by the institutional review board at Zhejiang University, Hangzhou, China. Animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee at the Zhejiang University, Hangzhou, China. Consent for publication: Not applicable. Conflict of interest: The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

