Characterization of gut microbiota and metabolites in renal transplant recipients during COVID-19 and prediction of one-year allograft function
- PMID: 40211390
- PMCID: PMC11987245
- DOI: 10.1186/s12967-025-06090-5
Characterization of gut microbiota and metabolites in renal transplant recipients during COVID-19 and prediction of one-year allograft function
Abstract
Background: The gut-lung-kidney axis is pivotal in immune-related kidney diseases, with gut dysbiosis potentially exacerbating the severity of Coronavirus disease 2019 (COVID-19) in recipients of kidney transplant. This study aimed to characterize the gut microbiome and metabolome in renal transplant recipients with COVID-19 pneumonia over a one-year follow-up period.
Methods: A total of 30 renal transplant recipients were enrolled, comprising 17 with COVID-19 pneumonia, six with mild COVID-19, and seven without COVID-19. Fecal samples were collected at the onset of infection for gut microbiome and metabolome analysis. Generalized Estimating Equations (GEE) model and Latent Class Growth Mixed Model (LCGMM) were employed to dissect the relationships among clinical characteristics, laboratory tests, and gut microbiota and metabolites.
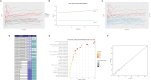
Results: Four microbial phyla (Deferribacteres, TM7, Fusobacteria, and Gemmatimonadetes) and 13 genera were significantly enriched across three recipients groups, correlating with baseline inflammatory response and allograft function. Additionally, 52 differentially expressed metabolites were identified, with seven significantly correlating with eight altered microbiota genera. LCGMM revealed two distinct classes of recipients, with those suffering from COVID-19 pneumonia exhibiting significantly elevated serum creatinine (Scr) trajectories over the one-year period. GEE further identified 12 genera and 181 metabolites closely associated with these trajectories; a multivariable model incorporating gut metabolites of 1-Caffeoylquinic Acid and PMK was found to effectively predict one-year allograft function.
Conclusions: Our study indicates a possible interaction between the composition of the gut microbiota and metabolites community and COVID-19 in renal transplant recipients, particularly in relation to disease severity and the prediction of one-year allograft function.
Keywords: Allograft function; COVID-19; Gut metabolome; Gut microbiome; Kidney transplantation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors of this manuscript have no conflicts of interest to disclose. Ethics approval and consent to participate: The study was conducted following the declaration of Helsinki. Ethical approval for this study was granted by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (2023-SRFA-007). Patient consent: Written informed consent from all transplant recipients was obtained. The study was strictly limited to the living-related transplantations of kidney donors to lineal or collateral relatives not beyond the third degree of kinship, or transplantations of kidney donors from cadaveric allograft donors after cardiac death.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

