Peptide Property Prediction for Mass Spectrometry Using AI: An Introduction to State of the Art Models
- PMID: 40211610
- PMCID: PMC12076536
- DOI: 10.1002/pmic.202400398
Peptide Property Prediction for Mass Spectrometry Using AI: An Introduction to State of the Art Models
Abstract
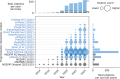
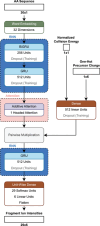
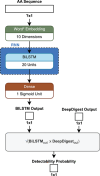
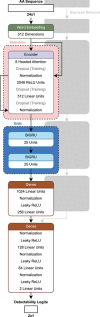
This review explores state of the art machine learning and deep learning models for peptide property prediction in mass spectrometry-based proteomics, including, but not limited to, models for predicting digestibility, retention time, charge state distribution, collisional cross section, fragmentation ion intensities, and detectability. The combination of these models enables not only the in silico generation of spectral libraries but also finds many additional use cases in the design of targeted assays or data-driven rescoring. This review serves as both an introduction for newcomers and an update for experienced researchers aiming to develop accessible and reproducible models for peptide property predictions. Key limitations of the current models, including difficulties in handling diverse post-translational modifications and instrument variability, highlight the need for large-scale, harmonized datasets, and standardized evaluation metrics for benchmarking.
Keywords: deep learning; machine learning; mass spectrometry; peptide property prediction; proteomics.
© 2025 The Author(s). Proteomics published by Wiley‐VCH GmbH.
Conflict of interest statement
M.W. is a founder and shareholder of OmicScouts GmbH and MSAID GmbH with no operational role in either company. The other authors do not have any conflicts of interest.
Figures







Similar articles
-
To Fly, or Not to Fly, That Is the Question: A Deep Learning Model for Peptide Detectability Prediction in Mass Spectrometry.J Proteome Res. 2025 Jun 6;24(6):2709-2726. doi: 10.1021/acs.jproteome.4c00973. Epub 2025 May 9. J Proteome Res. 2025. PMID: 40344201 Free PMC article.
-
Rescoring Peptide Spectrum Matches: Boosting Proteomics Performance by Integrating Peptide Property Predictors Into Peptide Identification.Mol Cell Proteomics. 2024 Jul;23(7):100798. doi: 10.1016/j.mcpro.2024.100798. Epub 2024 Jun 11. Mol Cell Proteomics. 2024. PMID: 38871251 Free PMC article. Review.
-
Machine learning-based peptide-spectrum match rescoring opens up the immunopeptidome.Proteomics. 2024 Apr;24(8):e2300336. doi: 10.1002/pmic.202300336. Epub 2023 Nov 27. Proteomics. 2024. PMID: 38009585 Review.
-
Ionmob: a Python package for prediction of peptide collisional cross-section values.Bioinformatics. 2023 Sep 2;39(9):btad486. doi: 10.1093/bioinformatics/btad486. Bioinformatics. 2023. PMID: 37540201 Free PMC article.
-
[Research progress and application of retention time prediction method based on deep learning].Se Pu. 2021 Mar;39(3):211-218. doi: 10.3724/SP.J.1123.2020.08015. Se Pu. 2021. PMID: 34227303 Free PMC article. Chinese.
References
-
- OpenAI, "ChatGPT," (2024), https://chatgpt.com/.
-
- stability.ai, "Image‐Models," (2024), https://stability.ai/stable‐image.
-
- Google, "Gemini," (2024), https://gemini.google.com/.
-
- Marr B., “Why Hybrid AI Is The Next Big Thing In Tech,” Forbes (2024), https://www.forbes.com/sites/bernardmarr/2024/10/02/why‐hybrid‐ai‐is‐the....
-
- Kleinman Z., “Microsoft: 'ever present' AI Assistants Are Coming,” BBC (2024), https://www.bbc.com/news/articles/czj9vmnlv9zo.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

