Role and mechanism of mitochondrial dysfunction‑related gene biomarkers in the progression of type 2 diabetes mellitus
- PMID: 40211718
- PMCID: PMC12004108
- DOI: 10.3892/mmr.2025.13523
Role and mechanism of mitochondrial dysfunction‑related gene biomarkers in the progression of type 2 diabetes mellitus
Abstract
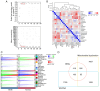
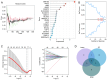
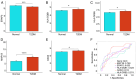
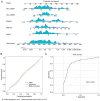
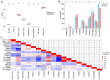
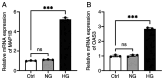
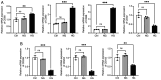
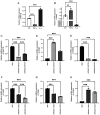
The present study aimed to elucidate the roles and mechanisms of gene biomarkers associated with mitochondrial dysfunction in the progression of Type 2 diabetes mellitus (T2DM). It conducted an analysis of differentially expressed genes related to mitochondrial dysfunction in T2DM and employed bioinformatics approaches to predict potential target drugs for key biomarkers. Additionally, the present study used the EPIC algorithm to examine immune cell infiltration in T2DM. Furthermore, the single‑cell RNA sequencing dataset GSE221156 was analyzed to identify specific cell types involved in T2DM. The expression of biomarkers was investigated through cellular experiments to assess the effect of marker genes on macrophage polarization. A total of five biomarker genes associated with T2DM were identified, namely ERAP2, HLA‑DQB1, HLA‑DRB5, MAP1B and OAS3. The combined detection of these genes yielded a risk‑predictive area under the curve value of 0.833 for T2DM. These five marker genes may serve as potential targets for valproic acid (VPA). During the progression of T2DM, there is an increase in macrophage numbers, with these genes being highly expressed in macrophages. In a high glucose‑induced RAW264.7 macrophage model, the expressions of MAP1B and OAS3 were upregulated. Notably, the knockdown of OAS3 markedly reduced M1 macrophage polarization, indicating OAS3 facilitates M1 macrophage polarization in a high‑glucose environment. The downregulation of OAS3 expression attenuated M1 macrophage polarization by inhibiting mTORC activation. In conclusion, five candidate biomarkers for T2DM were identified that may serve as therapeutic targets for VPA and are associated with immune infiltration in T2DM. Among these, OAS3 enhances M1 macrophage polarization in a high‑glucose environment by regulating the mTORC1 pathway.
Keywords: OAS3; macrophage polarization; mitochondrial dysfunction; type 2 diabetes; valproic acid.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Diane A, Allouch A, Mu UMRBA, Al-Siddiqi HH. Endoplasmic reticulum stress in pancreatic β-cell dysfunctionality and diabetes mellitus: A promising target for generation of functional hPSC-derived β-cells in vitro. Front Endocrinol (Lausanne) 2024;15:1386471. doi: 10.3389/fendo.2024.1386471. - DOI - PMC - PubMed
-
- Ding Y, Shi Y, Guan R, Yan S, Liu H, Wang Z, Li J, Wang T, Cai W, Ma G. Evaluation and comparison of efficacy and safety of tirzepatide and semaglutide in patients with type 2 diabetes mellitus: A Bayesian network meta-analysis. Pharmacol Res. 2024;199:107031. doi: 10.1016/j.phrs.2023.107031. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

