Influence of Sequencing Technology on Pangenome-Level Analysis and Detection of Antimicrobial Resistance Genes in ESKAPE Pathogens
- PMID: 40212029
- PMCID: PMC11983279
- DOI: 10.1093/ofid/ofaf183
Influence of Sequencing Technology on Pangenome-Level Analysis and Detection of Antimicrobial Resistance Genes in ESKAPE Pathogens
Abstract
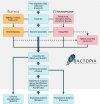
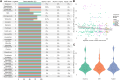
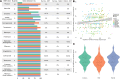
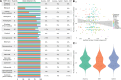
As sequencing costs decrease, short-read and long-read technologies are indispensable tools for uncovering the genetic drivers behind bacterial pathogen resistance. This study explores the differences between the use of short-read (Illumina) and long-read (Oxford Nanopore Technologies [ONT]) sequencing in detecting antimicrobial resistance (AMR) genes in ESKAPE pathogens (ie, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter cloacae). Utilizing a dataset of 1385 whole genome sequences and applying commonly used bioinformatic methods in bacterial genomics, we assessed the differences in genomic completeness, pangenome structure, and AMR gene and point mutation identification. Illumina presented higher genome completeness, while ONT identified a broader pangenome. Hybrid assembly outperformed both Illumina and ONT at identifying key AMR genetic determinants, presented results closer to Illumina's completeness, and revealed ONT-like pangenomic content. Notably, Illumina consistently detected more AMR-related point mutations than its counterparts. This highlights the importance of method selection based on research goals, particularly when using publicly available data ranging a wide timespan. Differences were also observed for specific gene classes and bacterial species, underscoring the need for a nuanced understanding of technology limitations. Overall, this study reveals the strengths and limitations of each approach, advocating for the use of Illumina for common AMR analysis, ONT for studying complex genomes and novel species, and hybrid assembly for a more comprehensive characterization, leveraging the benefits of both technologies.
Keywords: ESKAPE pathogens; Illumina; Oxford Nanopore Technologies; antimicrobial resistance; bacterial genomics.
© The Author(s) 2025. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. The authors: No reported conflicts of interest.
Figures



Update of
-
Influence of Sequencing Technology on Pangenome-level Analysis and Detection of Antimicrobial Resistance Genes in ESKAPE Pathogens.bioRxiv [Preprint]. 2025 Jan 10:2025.01.08.631980. doi: 10.1101/2025.01.08.631980. bioRxiv. 2025. Update in: Open Forum Infect Dis. 2025 Mar 26;12(4):ofaf183. doi: 10.1093/ofid/ofaf183. PMID: 39829834 Free PMC article. Updated. Preprint.
Similar articles
-
Influence of Sequencing Technology on Pangenome-level Analysis and Detection of Antimicrobial Resistance Genes in ESKAPE Pathogens.bioRxiv [Preprint]. 2025 Jan 10:2025.01.08.631980. doi: 10.1101/2025.01.08.631980. bioRxiv. 2025. Update in: Open Forum Infect Dis. 2025 Mar 26;12(4):ofaf183. doi: 10.1093/ofid/ofaf183. PMID: 39829834 Free PMC article. Updated. Preprint.
-
Evaluation of DNA extraction kits for long-read shotgun metagenomics using Oxford Nanopore sequencing for rapid taxonomic and antimicrobial resistance detection.Sci Rep. 2024 Nov 27;14(1):29531. doi: 10.1038/s41598-024-80660-3. Sci Rep. 2024. PMID: 39604411 Free PMC article.
-
Three Distinct Annotation Platforms Differ in Detection of Antimicrobial Resistance Genes in Long-Read, Short-Read, and Hybrid Sequences Derived from Total Genomic DNA or from Purified Plasmid DNA.Antibiotics (Basel). 2022 Oct 12;11(10):1400. doi: 10.3390/antibiotics11101400. Antibiotics (Basel). 2022. PMID: 36290058 Free PMC article.
-
Unseen Enemy: Mechanisms of Multidrug Antimicrobial Resistance in Gram-Negative ESKAPE Pathogens.Antibiotics (Basel). 2025 Jan 9;14(1):63. doi: 10.3390/antibiotics14010063. Antibiotics (Basel). 2025. PMID: 39858349 Free PMC article. Review.
-
Metal Nanoparticle-Based Biosensors for the Early Diagnosis of Infectious Diseases Caused by ESKAPE Pathogens in the Fight against the Antimicrobial-Resistance Crisis.Biosensors (Basel). 2024 Jul 11;14(7):339. doi: 10.3390/bios14070339. Biosensors (Basel). 2024. PMID: 39056615 Free PMC article. Review.
References
-
- Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. Chicago, IL: University of Chicago Press, 2008:1079–81. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

