A Novel Piezo1 Agonist Promoting Mesenchymal Stem Cell Proliferation and Osteogenesis to Attenuate Disuse Osteoporosis
- PMID: 40212079
- PMCID: PMC11935128
- DOI: 10.1002/smsc.202400061
A Novel Piezo1 Agonist Promoting Mesenchymal Stem Cell Proliferation and Osteogenesis to Attenuate Disuse Osteoporosis
Abstract
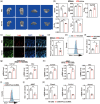
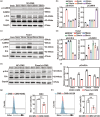
Disuse osteoporosis (OP) is a state of bone loss due to lack of mechanical stimuli, probably induced by prolonged bed rest, neurological diseases, as well as microgravity. Currently the precise treatment strategies of disuse OP remain largely unexplored. Piezo1, a mechanosensitive calcium (Ca2+) ion channel, is a key force sensor mediating mechanotransduction and it is demonstrated to regulate bone homeostasis and osteogenesis in response to mechanical forces. Using structure-based drug design, a novel small-molecule Piezo1 agonist, MCB-22-174, which can effectively activate Piezo1 and initiate Ca2+ influx, is developed and is more potent than the canonical Piezo1 agonist, Yoda1. Moreover, MCB-22-174 is found as a safe Piezo1 agonist without any signs of serious toxicity. Mechanistically, Piezo1 activation promotes the proliferation of bone marrow mesenchymal stem cells by activating the Ca2+-related extracellular signal-related kinases and calcium-calmodulin (CaM)-dependent protein kinase II (CaMKII) pathway. Importantly, MCB-22-174 could effectively promote osteogenesis and attenuate disuse OP in vivo. Overall, the findings provide a promising therapeutic strategy for disuse OP by chemical activation of Piezo1.
Keywords: Piezo1; disuse osteoporosis; mechenchymal stem cell proliferations; novel agonists; osteogenesis.
© 2024 The Author(s). Small Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Watanabe‐Takano H., Ochi H., Chiba A., Matsuo A., Kanai Y., Fukuhara S., Ito N., Sako K., Miyazaki T., Tainaka K., Harada I., Sato S., Sawada Y., Minamino N., Takeda S., Ueda H. R., Yasoda A., Mochizuki N., Cell Rep. 2021, 36, 109380. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
