Identification of Gene Targets for the Sprouting Inhibitor CIPC
- PMID: 40212537
- PMCID: PMC11982522
- DOI: 10.1002/pld3.70068
Identification of Gene Targets for the Sprouting Inhibitor CIPC
Abstract
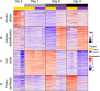
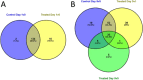
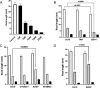
Sprout suppressants are widely used in industry to ensure year-round availability of potato tubers, significantly decreasing wastage by repressing premature growth of buds on the tuber surface during storage. Despite its ban from 2020 in the EU, isopropyl N-(3-chlorophenyl) carbamate (also known as chlorpropham or CIPC) remains the most widely used suppressant worldwide. However, the mechanism of action of CIPC remains obscure. Here, we report on a combined targeted transcriptomic and genetic approach to identify components in the tuber bud cell-division machinery that might be involved in CIPC's mode of action. This involved RNAseq analysis of dissected, staged tuber buds during in vitro sprouting with and without CIPC to identify lead genes, followed by the development and application of an Arabidopsis root assay to assess cell division response to CIPC in selected mutants. The ease of use of this model plant, coupled with its immense genetic resources, allowed us to test the functionality of lead genes encoding cell-division-associated proteins in the modulation of plant growth response to CIPC. This approach led to the identification of a component of the augmin complex (a core player in mitosis) as a potential target for CIPC.
Keywords: CIPC; cell division; chlorpropham; potato; sprouting.
© 2025 The Author(s). Plant Direct published by American Society of Plant Biologists and the Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
The sprout inhibitors chlorpropham and 1,4-dimethylnaphthalene elicit different transcriptional profiles and do not suppress growth through a prolongation of the dormant state.Plant Mol Biol. 2010 May;73(1-2):181-9. doi: 10.1007/s11103-010-9607-6. Epub 2010 Feb 5. Plant Mol Biol. 2010. PMID: 20135197
-
Sprout suppression on potato: need to look beyond CIPC for more effective and safer alternatives.J Food Sci Technol. 2016 Jan;53(1):1-18. doi: 10.1007/s13197-015-1980-3. Epub 2015 Aug 13. J Food Sci Technol. 2016. PMID: 26787928 Free PMC article. Review.
-
Biodegradation of chlorpropham and its major products by Bacillus licheniformis NKC-1.World J Microbiol Biotechnol. 2018 Jul 6;34(8):112. doi: 10.1007/s11274-018-2494-8. World J Microbiol Biotechnol. 2018. PMID: 29980862
-
Mint essential oil can induce or inhibit potato sprouting by differential alteration of apical meristem.Planta. 2010 Jun;232(1):179-86. doi: 10.1007/s00425-010-1154-5. Planta. 2010. PMID: 20390295
-
Assuring Potato Tuber Quality during Storage: A Future Perspective.Front Plant Sci. 2017 Nov 28;8:2034. doi: 10.3389/fpls.2017.02034. eCollection 2017. Front Plant Sci. 2017. PMID: 29234341 Free PMC article. Review.
References
-
- Agriculture and Horticulture Development Board 2021. (DMN) 1,4‐dimethylnaphthalene. https://ahdb.org.uk/sprout‐suppression‐series‐1‐4‐dimethylnaphthalene‐dmn.
-
- Bartels, P. G. , and Hilton J. L.. 1973. “Comparison of Trifluralin, Oryzalin, Pronamide, Propham, and Colchicine Treatments on Microtubules.” Pesticide Biochemistry and Physiology 3, no. 4: 462–472. 10.1016/0048-3575(73)90072-2. - DOI
-
- Campbell, M. A. , Gleichsner A., Alsbury R., Horvath D., and Suttle J.. 2010. “The Sprout Inhibitors Chlorpropham and 1,4‐Dimethylnaphthalene Elicit Different Transcriptional Profiles and Do Not Suppress Growth Through a Prolongation of the Dormant State.” Plant Molecular Biology 73, no. 1–2: 181–189. 10.1007/s11103-010-9607-6. - DOI - PubMed
-
- Dewitte, W. , Scofield S., Alcasabas A. A., et al. 2007. “Arabidopsis CYCD3 D‐Type Cyclins Link Cell Proliferation and Endocycles and Are Rate‐Limiting for Cytokinin Responses.” Proceedings of the National Academy of Sciences of the United States of America 104, no. 36: 14537–14542. 10.1073/pnas.0704166104. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

