Genome assembly and annotation of Acropora pulchra from Mo'orea French Polynesia
- PMID: 40212849
- PMCID: PMC11985253
- DOI: 10.46471/gigabyte.153
Genome assembly and annotation of Acropora pulchra from Mo'orea French Polynesia
Abstract
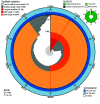
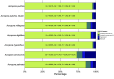
Reef-building corals are integral ecosystem engineers of tropical reefs but face threats from climate change. Investigating genetic, epigenetic, and environmental factors influencing their adaptation is critical. Genomic resources are essential for understanding coral biology and guiding conservation efforts. However, genomes of the coral genus Acropora are limited to highly-studied species. Here, we present the assembly and annotation of the genome and DNA methylome of Acropora pulchra from Mo'orea, French Polynesia. Using long-read PacBio HiFi and Illumina RNASeq, we generated the most complete Acropora genome to date (BUSCO completeness of 96.7% metazoan genes). The assembly size is 518 Mbp, with 174 scaffolds, and a scaffold N50 of 17 Mbp. We predicted 40,518 protein-coding genes and 16.74% of the genome in repeats. DNA methylation in the CpG context is 14.6%. This assembly of the A. pulchra genome and DNA methylome will support studies of coastal corals in French Polynesia, aiding conservation and comparative studies of Acropora and cnidarians.
© The Author(s) 2025.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- van Oppen MJH, McDonald BJ, Willis B et al. . The evolutionary history of the coral Genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: reticulation, incomplete lineage sorting, or morphological convergence? Mol. Biol. Evol., 2001; 18(7): 1315–1329. doi:10.1093/oxfordjournals.molbev.a003916. - DOI - PubMed
-
- Souter P, Willis BL, Bay LK et al. . Location and disturbance affect population genetic structure in four coral species of the genus Acropora on the Great Barrier Reef. Mar. Ecol. Prog. Ser., 2010; 416: 35–45. doi:10.3354/meps08740. - DOI
-
- Rios D. . The population genetic structure of Acropora pulchra in Guam. Thesis, University of Guam, 2020. https://gepscor-cdn.uog.edu/wp-content/uploads/2023/05/MLThesis_RiosD.pdf.
LinkOut - more resources
Full Text Sources
Miscellaneous
