Plant stem and leaf segmentation and phenotypic parameter extraction using neural radiance fields and lightweight point cloud segmentation networks
- PMID: 40212877
- PMCID: PMC11983422
- DOI: 10.3389/fpls.2025.1491170
Plant stem and leaf segmentation and phenotypic parameter extraction using neural radiance fields and lightweight point cloud segmentation networks
Abstract
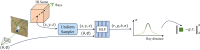
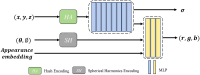
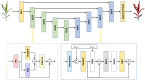
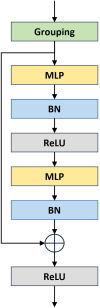
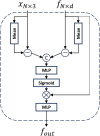
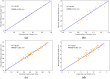
High-quality 3D reconstruction and accurate 3D organ segmentation of plants are crucial prerequisites for automatically extracting phenotypic traits. In this study, we first extract a dense point cloud from implicit representations, which derives from reconstructing the maize plants in 3D by using the Nerfacto neural radiance field model. Second, we propose a lightweight point cloud segmentation network (PointSegNet) specifically for stem and leaf segmentation. This network includes a Global-Local Set Abstraction (GLSA) module to integrate local and global features and an Edge-Aware Feature Propagation (EAFP) module to enhance edge-awareness. Experimental results show that our PointSegNet achieves impressive performance compared to five other state-of-the-art deep learning networks, reaching 93.73%, 97.25%, 96.21%, and 96.73% in terms of mean Intersection over Union (mIoU), precision, recall, and F1-score, respectively. Even when dealing with tomato and soybean plants, with complex structures, our PointSegNet also achieves the best metrics. Meanwhile, based on the principal component analysis (PCA), we further optimize the method to obtain the parameters such as leaf length and leaf width by using PCA principal vectors. Finally, the maize stem thickness, stem height, leaf length, and leaf width obtained from our measurements are compared with the manual test results, yielding R 2 values of 0.99, 0.84, 0.94, and 0.87, respectively. These results indicate that our method has high accuracy and reliability for phenotypic parameter extraction. This study throughout the entire process from 3D reconstruction of maize plants to point cloud segmentation and phenotypic parameter extraction, provides a reliable and objective method for acquiring plant phenotypic parameters and will boost plant phenotypic development in smart agriculture.
Keywords: lightweight network; neural radiance fields; plant phenotype; point cloud segmentation; three-dimensional point cloud.
Copyright © 2025 Qiao, Zhang, Niu, Han and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
DFSP: A fast and automatic distance field-based stem-leaf segmentation pipeline for point cloud of maize shoot.Front Plant Sci. 2023 Jan 31;14:1109314. doi: 10.3389/fpls.2023.1109314. eCollection 2023. Front Plant Sci. 2023. PMID: 36798707 Free PMC article.
-
Automatic Branch-Leaf Segmentation and Leaf Phenotypic Parameter Estimation of Pear Trees Based on Three-Dimensional Point Clouds.Sensors (Basel). 2023 May 8;23(9):4572. doi: 10.3390/s23094572. Sensors (Basel). 2023. PMID: 37177776 Free PMC article.
-
A cotton organ segmentation method with phenotypic measurements from a point cloud using a transformer.Plant Methods. 2025 Mar 16;21(1):37. doi: 10.1186/s13007-025-01357-w. Plant Methods. 2025. PMID: 40091062 Free PMC article.
-
Measurement of Maize Leaf Phenotypic Parameters Based on 3D Point Cloud.Sensors (Basel). 2025 Apr 30;25(9):2854. doi: 10.3390/s25092854. Sensors (Basel). 2025. PMID: 40363288 Free PMC article.
-
Three-dimensional branch segmentation and phenotype extraction of maize tassel based on deep learning.Plant Methods. 2023 Aug 1;19(1):76. doi: 10.1186/s13007-023-01051-9. Plant Methods. 2023. PMID: 37528454 Free PMC article.
References
-
- Deng X., Zhang W., Ding Q., Zhang X. (2023). “Pointvector: a vector representation in point cloud analysis,” in 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada. (New York: IEEE; ), 9455–9465.
-
- Guo M.-H., Cai J.-X., Liu Z.-N., Mu T.-J., Martin R. R., Hu S.-M. (2021). Pct: Point cloud transformer. Comput. Visual Media 7, 187–199. doi: 10.1007/s41095-021-0229-5 - DOI
-
- Guo Q., Jin S., Li M., Yang Q., Xu K., Ju Y., et al. . (2020). Application of deep learning in ecological resource research: Theories, methods, and challenges. Sci. China Earth Sci. 63, 1457–1474. doi: 10.1007/s11430-019-9584-9 - DOI
LinkOut - more resources
Full Text Sources

