Hepatic stellate cell-derived microfibrillar-associated protein 2 prevents liver fibrosis by regulating extracellular matrix and inflammation
- PMID: 40213670
- PMCID: PMC11980660
- DOI: 10.7150/thno.109771
Hepatic stellate cell-derived microfibrillar-associated protein 2 prevents liver fibrosis by regulating extracellular matrix and inflammation
Abstract
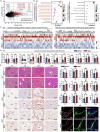
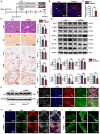
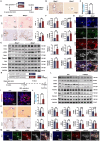
Microfibrillar-associated protein 2 (MFAP-2) is a crucial component of the extracellular matrix (ECM) microfibrils, yet its role in liver fibrosis remains elusive. Methods: Human tissue arrays and mouse models of fibrosis progression and resolution were used to investigate MFAP-2 expression patterns. Mfap2 deficiency (Mfap2 -/-) or overexpression (ovMfap2) mice were subjected to carbon tetrachloride (CCl4) injection or bile duct ligation (BDL) to induce liver fibrosis. Histological, biochemical, bulk, or single-cell RNA-sequencing (scRNA-seq), proteomics to analyze the matrisome, and in vitro studies were conducted. Results: MFAP-2 was predominantly enriched in activated hepatic stellate cells (HSCs) and upregulated in advanced liver fibrosis. Although Mfap2 ablation had minimal impact on collagen deposition during CCl4 injection, it significantly delayed fibrosis regression after CCl4 cessation. The delayed fibrosis regression due to Mfap2 deficiency was likely linked to aggravated intrahepatic inflammation, ECM stabilization, and activated focal adhesion signaling in HSCs. Mechanically, inhibiting HSC-derived Mfap2 enhanced HSC interactions and increased matrisome protein production, while reducing the interaction between HSCs and liver-resident macrophages by decreasing macrophage migration inhibitory factor secretion from HSCs. Additionally, we validated the role of Mfap2 deletion in liver fibrosis using the BDL mouse model, demonstrating a more pronounced effect on fibrosis progression. Adeno-associated virus vector (serotype 6)-mediated Mfap2 overexpression in HSCs conferred protection against liver fibrosis in both models. Conclusion: This study reveals the compensatory protective effects of HSC-derived MFAP-2 on liver fibrosis and its underlying mechanisms. Enhancing MFAP-2 in HSCs may therefore benefit patients with liver fibrosis.
Keywords: Liver fibrosis; extracellular matrix; hepatic stellate cell; macrophage; matrisome.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Sun Y, Chen W, Chen S, Wu X, Zhang X, Zhang L. et al. Regression of Liver Fibrosis in Patients on Hepatitis B Therapy Is Associated With Decreased Liver-Related Events. Clin Gastroenterol Hepatol. 2024;22:591–601. e3. - PubMed
-
- Sun Y, Zhou J, Wang L, Wu X, Chen Y, Piao H. et al. New classification of liver biopsy assessment for fibrosis in chronic hepatitis B patients before and after treatment. Hepatology. 2017;65:1438–50. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

