Targets of the transcription factor Six1 identify previously unreported candidate deafness genes
- PMID: 40213817
- PMCID: PMC12045605
- DOI: 10.1242/dev.204533
Targets of the transcription factor Six1 identify previously unreported candidate deafness genes
Abstract
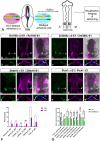
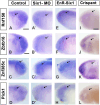
Branchio-otic (BOS) and branchio-oto-renal (BOR) syndromes are autosomal dominant disorders featuring multiple birth defects including ear, renal and branchial malformations. Mutations in the homeodomain transcription factor SIX1 and its co-factor EYA1 have been identified in about 50% of individuals with BOS or BOR, while causative mutations are unknown in the other half. We hypothesise that SIX1 target genes represent new BOS and BOR candidates. Using published transcriptomic and epigenomic data from chick ear progenitors, we first identify putative Six1 targets. Next, we provide evidence that Six1 directly regulates some of these candidates: Six1 binds to their enhancers, and functional experiments in Xenopus and chick confirm that Six1 controls their expression. Finally, we show that most putative chick Six1 targets are also expressed in the human developing ear and are associated with known deafness loci. Together, our results not only characterise the molecular mechanisms that mediate Six1 function in the developing ear, but also provide new candidates for human congenital deafness.
Keywords: Branchio-oto-renal Syndrome; Deafness; Ear development; Otic placode.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



Similar articles
-
EYA1 mutations associated with the branchio-oto-renal syndrome result in defective otic development in Xenopus laevis.Biol Cell. 2010 Feb 17;102(5):277-92. doi: 10.1042/BC20090098. Biol Cell. 2010. PMID: 19951260 Free PMC article.
-
Six1 proteins with human branchio-oto-renal mutations differentially affect cranial gene expression and otic development.Dis Model Mech. 2020 Mar 3;13(3):dmm043489. doi: 10.1242/dmm.043489. Dis Model Mech. 2020. PMID: 31980437 Free PMC article.
-
Genetic research progress in branchio-oto syndrome/ branchio-oto-renal syndrome.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2022 Jan 28;47(1):129-138. doi: 10.11817/j.issn.1672-7347.2022.210251. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2022. PMID: 35545373 Free PMC article. Chinese, English.
-
Branchio-oto-renal syndrome: comprehensive review based on nationwide surveillance in Japan.Pediatr Int. 2014 Jun;56(3):309-14. doi: 10.1111/ped.12357. Pediatr Int. 2014. PMID: 24730701 Review.
-
Branchio-oto-renal syndrome.Am J Med Genet A. 2007 Jul 15;143A(14):1671-8. doi: 10.1002/ajmg.a.31561. Am J Med Genet A. 2007. PMID: 17238186 Review.
Cited by
-
Sponge bHLH Gene Expression in Xenopus laevis Disrupts Inner Ear and Lateral Line Neurosensory Development and Otic Afferent Pathfinding.Int J Mol Sci. 2025 Jun 7;26(12):5487. doi: 10.3390/ijms26125487. Int J Mol Sci. 2025. PMID: 40564948 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

