DNA Molecular Computing with Weighted Signal Amplification for Cancer miRNA Biomarker Diagnostics
- PMID: 40213938
- PMCID: PMC12165086
- DOI: 10.1002/advs.202416490
DNA Molecular Computing with Weighted Signal Amplification for Cancer miRNA Biomarker Diagnostics
Abstract
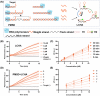
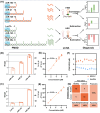
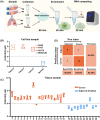
The expression levels of microRNAs (miRNAs) are strongly linked to cancer progression, making them promising biomarkers for cancer detection. Enzyme-free signal amplification DNA circuits have facilitated the detection of low-abundance miRNAs. However, these methods may neglect the diagnostic value (or weight) of different miRNAs. Here, a molecular computing approach with weighted signal amplification is presented. Polymerase-mediated strand displacement is employed to assign weights to target miRNAs, reflecting the miRNAs' diagnostic values, followed by amplification of the weighted signals using localized DNA catalytic hairpin assembly. This method is applied to diagnose miRNAs for non-small cell lung cancer (NSCLC). Machine learning is used to identify NSCLC-specific miRNAs and assign corresponding weights for optimum classification of healthy and lung cancer individuals. With the molecular computing of the miRNAs, the diagnostic output is simplified as a single channel of fluorescence intensity. Cancer tissues (n = 18) and adjacent cancer tissues (n = 10) are successfully classified within 2.5 h (sample-to-result) with an accuracy of 92.86%. The weighted amplification strategy has the potential to extend to the digital detection of multidimensional biomarkers, advancing personalized disease diagnostics in point-of-care settings.
Keywords: DNA computing; NSCLC diagnostics; machine learning; miRNA; signal amplification.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Detection of NSCLC biomarker miRNAs via localized catalytic hairpin self-assembly and laser-induced fluorescence on a microfluidic platform.Analyst. 2025 May 12;150(10):2160-2169. doi: 10.1039/d5an00073d. Analyst. 2025. PMID: 40227732
-
MicroRNAs as novel biomarkers in the diagnosis of non-small cell lung cancer: a meta-analysis based on 20 studies.Tumour Biol. 2014 Sep;35(9):9119-29. doi: 10.1007/s13277-014-2188-2. Epub 2014 Jun 12. Tumour Biol. 2014. PMID: 25027397
-
MicroRNA-based biomarkers for diagnosis of non-small cell lung cancer (NSCLC).Thorac Cancer. 2020 Mar;11(3):762-768. doi: 10.1111/1759-7714.13337. Epub 2020 Jan 28. Thorac Cancer. 2020. PMID: 31994346 Free PMC article.
-
MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer.Cell Prolif. 2017 Dec;50(6):e12394. doi: 10.1111/cpr.12394. Epub 2017 Oct 8. Cell Prolif. 2017. PMID: 28990243 Free PMC article. Review.
-
Recent strategies for electrochemical sensing detection of miRNAs in lung cancer.Anal Biochem. 2023 Jan 15;661:114986. doi: 10.1016/j.ab.2022.114986. Epub 2022 Nov 13. Anal Biochem. 2023. PMID: 36384188 Review.
Cited by
-
Advances in Research on Isothermal Signal Amplification Mediated MicroRNA Detection of Clinical Samples: Application to Disease Diagnosis.Biosensors (Basel). 2025 Jun 18;15(6):395. doi: 10.3390/bios15060395. Biosensors (Basel). 2025. PMID: 40558477 Free PMC article. Review.
References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA Cancer J. Clin. 2021, 71, 209. - PubMed
-
- Wu Y. L., Dziadziuszko R., Ahn J. S., Barlesi F., Nishio M., Lee D. H., Lee J. S., Zhong W., Horinouchi H., Mao W., Hochmair M., de Marinis F., Migliorino M. R., Bondarenko I., Lu S., Wang Q., Ochi Lohmann T., Xu T., Cardona A., Ruf T., Noe J., Solomon B. J., N. Engl. J. Med. 2024, 390, 1265. - PubMed
-
- Liu C., Zhao J., Tian F., Cai L., Zhang W., Feng Q., Chang J., Wan F., Yang Y., Dai B., Cong Y., Ding B., Sun J., Tan W., Nat. Biomed. Eng. 2019, 3, 183. - PubMed
-
- Stinchcombe T. E., Socinski M. A., Nat. Rev. Clin. Oncol. 2010, 7, 426. - PubMed
-
- Camidge D. R., Doebele R. C., Kerr K. M., Nat. Rev. Clin. Oncol. 2019, 16, 341. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
