Episomal virus maintenance enables bacterial population recovery from infection and promotes virus-bacterial coexistence
- PMID: 40214158
- PMCID: PMC12064560
- DOI: 10.1093/ismejo/wraf066
Episomal virus maintenance enables bacterial population recovery from infection and promotes virus-bacterial coexistence
Abstract
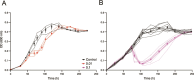
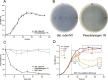
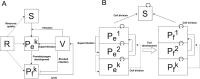
Viruses are ubiquitous in aquatic environments with total densities of virus-like particles often exceeding 107/ml in surface marine oligotrophic waters. Hypersaline environments harbor elevated prokaryotic population densities of 108/ml that coexist with viruses at even higher densities, approaching 1010/ml. The presence of high densities of microbial populations and viruses challenge traditional explanations of top-down control exerted by viruses. At close to saturation salinities, prokaryotic populations are dominated by Archaea and the bacterial genus Salinibacter. In this work we examine the episomal maintenance of a virus within a Salinibacter ruber host. We found that infected cultures of Sal. ruber M1 developed a population-level resistance and underwent systematic and reproducible recovery post infection that was counter-intuitively dependent on the multiplicity of infection, where higher viral pressures led to better host outcomes. Furthermore, we developed a nonlinear population dynamics model that successfully reproduced the qualitative features of the recovery. Together, experiments and models suggest that episomal virus maintenance and lysis inhibition enable host-virus co-existence at high viral densities. Our results emphasize the ecological importance of exploring a spectrum of viral infection strategies beyond the conventional binary of lysis or lysogeny.
Keywords: Salinibacter; acquired phage resistance; modeling; pseudolysogeny; virus–host interactions.
© The Author(s) 2025. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
None declared.
Figures

References
-
- Shiklomanov IA. World Water Resources: A New Appraisal and Assessment for the 21st Century. Paris: UNESCO-IHP, 1998.
-
- Ventosa A. Unusual Micro-Organisms from Unusual Habitats: Hypersaline Environments. Cambridge: Cambridge University Press, 2006.. 10.1017/CBO9780511754913.015 223 54. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

