Toward a deeper understanding of dengue: novel method for quantification and isolation of envelope protein epitope-specific antibodies
- PMID: 40214258
- PMCID: PMC12108060
- DOI: 10.1128/msphere.00961-24
Toward a deeper understanding of dengue: novel method for quantification and isolation of envelope protein epitope-specific antibodies
Abstract
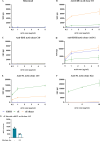
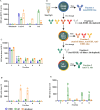
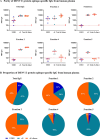
The dengue viruses (DENV) envelope (E) protein is the main target of the antibody (Ab) response. Abs target different epitopes on the E-protein, including sE-dimer, E domain III (EDIII), and fusion loop (FL). Anti-EDIII Abs are mainly serotype-specific, whereas anti-FL Abs can induce antibody-dependent enhancement (ADE) in vitro. Abs targeting sE-dimer epitopes can cross-neutralize different DENV serotypes. However, the involvement of each Ab subset in disease pathogenicity and/or protection remains unclear. We aimed to optimize the quantification and purification of DENV E-protein epitope-specific Abs from human samples. C-terminal biotinylated DENV2 E recombinant proteins (EDIII, soluble E [sE], and sE-dimer) were coupled to color-coded magnetic microspheres for a multiplex immunoassay (MIA), testing different antigen concentrations. Assay performance was evaluated using well-characterized anti-DENV monoclonal antibodies (mAbs) and total IgG from DENV seronegative and seropositive human plasma. Specific FL epitopes were blocked with mouse mAb clone 4G2 to quantify anti-FL- and sE-dimer-specific Abs, measuring antigen-antibody reactions as median fluorescence intensity (MFI). For isolation of E-protein epitope-specific antibodies, sE-proteins were conjugated to streptavidin resin beads. Total IgG from human plasma was incubated with immobilized EDIII to elute anti-EDIII Abs. The flow-through was incubated with sE-dimer resin beads to elute sE-dimer specific Ab enriched fraction, and the flow-through was applied to immobilized sE to elute anti-FL Abs. In conclusion, we have developed a serological assay to detect E-protein epitope-specific Abs in DENV-infected humans. Additionally, we successfully isolated anti-EDIII, anti-FL, and an enriched fraction of sE-dimer specific Abs from human samples.IMPORTANCEThe development of effective dengue virus (DENV) vaccines has been hampered by limited insights into the immunological mechanisms of protection. Our study addresses this gap by introducing a refined multiplex microsphere-based immunoassay (MIA) to quantify and isolate antibodies (Abs) targeting specific E-protein epitopes, such as E domain III (EDIII), the fusion loop (FL), and the sE-dimer specific Abs. This method provides detailed epitope-specific Ab profiling with high sensitivity and requires minimal sample volumes. The ability to isolate specific Ab subsets from human plasma also enables detailed investigations into their roles in protection or pathogenesis, paving the way for more effective dengue interventions.
Keywords: antibody response to dengue; antibody-dependent enhancement; dengue virus; flavivirus anti-envelope antibodies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Murugesan A, Manoharan M. 2020. Dengue virus, p 281–359. In Emerging and reemerging viral pathogens. 10.1016/B978-0-12-819400-3.00016-8. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

