Identification of Nuclear Localization Sequence (NLS) Sites in R2R3-MYB Transcription Factor Involved in Anther Development
- PMID: 40214424
- PMCID: PMC11987959
- DOI: 10.3390/cells14070470
Identification of Nuclear Localization Sequence (NLS) Sites in R2R3-MYB Transcription Factor Involved in Anther Development
Abstract
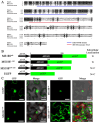
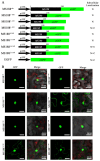
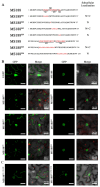
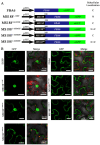
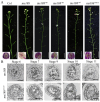
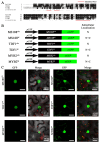
The R2R3-MYB family of transcription factors (TFs) plays a crucial role in cell specification and secondary metabolism regulation during plant development. In Arabidopsis, MS188, a typical R2R3-MYB protein, is essential for tapetal development and pollen wall formation. However, the nuclear localization sequence (NLS) responsible for directing MS188 into the nucleus has not been fully elucidated. In this study, the subcellular localization of the NLS-containing proteins was determined by GFP tagging in tobacco leaves, and three NLS regions within MS188 were identified: two located at the N-terminus of R2-MYB and one at the C-terminus of R3-MYB. We further narrowed the NLSs located at amino acids (AAs) 12-15, 18-22, and 96-107 via point mutation analysis. Combined with the cytoplasmic protein FBA6, these NLSs fusion proteins could localize in the nucleus. Importantly, the proteins with mutations in AAs 18-22 exhibited completely cytoplasmic signals, whereas other mutated sites partially abolished the nuclear signals. These findings suggest that the NLS at AAs 18-22 is sufficient for nuclear localization. To confirm the NLS functions in vivo, we constructed the vectors including the MS188 gene without the NLS sites, which failed to complement the male sterile phenotype of ms188. We also searched the highly conserved NLSs in other R2R3-MYB TFs and showed they are required for nuclear localization. Collectively, these findings revealed the specific NLS regions within R2R3-MYB transcription factors and highlighted their critical role for subcellular localization in plant developmental regulation.
Keywords: R2R3-MYB transcription factor; anther development; nuclear localization sequence.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
References
-
- Latchman D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997;29:1305–1312. - PubMed
-
- Amoutzias G.D., Robertson D.L., Van de Peer Y., Oliver S.G. Choose your partners: Dimerization in eukaryotic transcription factors. Trends Biochem. Sci. 2008;33:220–229. - PubMed
-
- Klempnauer K.-H., Gonda T.J., Michael Bishop J. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell. 1982;31:453–463. - PubMed
-
- Rosinski J.A., Atchley W.R. Molecular Evolution of the Myb Family of Transcription Factors: Evidence for Polyphyletic Origin. J. Mol. Evol. 1998;46:74–83. - PubMed
-
- Ogata K., Kanei-Ishii C., Sasaki M., Hatanaka H., Nagadoi A., Enari M., Nakamura H., Nishimura Y., Ishii S., Sarai A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 1996;3:178–187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

