TPM4 influences the initiation and progression of gastric cancer by modulating ferroptosis via SCD1
- PMID: 40214825
- PMCID: PMC11991984
- DOI: 10.1007/s10238-025-01629-8
TPM4 influences the initiation and progression of gastric cancer by modulating ferroptosis via SCD1
Abstract
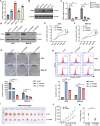
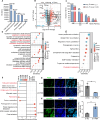
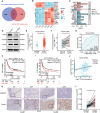
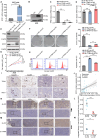
Gastric cancer (GC) is a deadly disease with poor prognosis and few treatment options. Tropomyosin 4 (TPM4) is an actin-binding protein that stabilizes the cytoskeleton of cells and has an unclear role in GC. This study aimed to elucidate the role and underlying mechanisms of TPM4 in GC pathogenesis. The expression and diagnostic and prognostic value of TPM4 in GC were analyzed using bioinformatics. A nomogram based on TPM4 expression was created and validated with an external cohort. TPM4-knockdown GC cells and xenograft models in nude mice were used to study the function of TPM4 in vitro and in vivo. Proteomic and rescue experiments confirmed the regulatory effect of TPM4 on stearoyl-CoA desaturase 1 (SCD1) in GC. Immunohistochemistry verified the expression and correlation of the TPM4 and SCD1 proteins in GC tissues. Our study identified TPM4 as an oncogene in GC, suggesting its potential diagnostic and prognostic value. The TPM4-based nomogram showed potential prognostic value for clinical use. TPM4 knockdown inhibited GC cell proliferation, induced ferroptosis, and slowed tumor growth in vivo, which is achieved by inhibiting SCD1 expression. Immunohistochemical analysis of GC tissues revealed elevated expression levels of both TPM4 and SCD1 proteins, with a positive correlation observed between their expression. TPM4 is a promising target for new diagnostic, prognostic, and therapeutic strategies for GC. Downregulation of TPM4 inhibits GC cell growth and induces ferroptosis by suppressing SCD1 expression.
Keywords: Ferroptosis; Gastric cancer; Proliferation; Stearoyl-CoA desaturase 1; Tropomyosin 4.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors state that they possess no conflicting interests related to the material presented in this article. Ethical approval: The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee at Qinghai University Affiliated Hospital (SL-2021128). The animal studies were approved by the animal ethics committee of Qinghai University (SL-2021128). Consent to participate: All tumor specimens analyzed in this study were formalin-fixed and paraffin-embedded, which had been confirmed as GC by specialist pathologist, and written consent for medical research purposes was obtained from the patients. Tissue sample collection was approved by the Ethics Committee at Qinghai University Affiliated Hospital (SL-2021128, date of sample collection approval: 2024.05).
Figures



References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660. - PubMed
-
- Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–64. 10.1016/S0140-6736(16)30354-3. - PubMed
-
- Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687–97. 10.1016/S0140-6736(10)61121-X. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

