The cohesin-associated protein Pds5A governs the meiotic spindle assembly via deubiquitination of Kif5B in oocytes
- PMID: 40215310
- PMCID: PMC11988415
- DOI: 10.1126/sciadv.adt6159
The cohesin-associated protein Pds5A governs the meiotic spindle assembly via deubiquitination of Kif5B in oocytes
Abstract
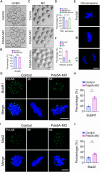
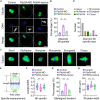
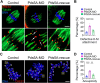
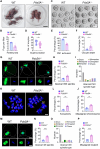
Chromosome cohesion mediated by cohesin complex and its associated proteins is required for accurate chromosome segregation and genomic stability in mitosis. However, because of the distinct operation mechanisms, many proteins might exert different functions during meiosis in germ cells. Here, we document that cohesin-associated protein precocious dissociation of sisters 5A (Pds5A) plays a noncanonical role in the meiotic spindle assembly during oocyte maturation independent of its cohesion function. Pds5A distributes on the spindle fibers in oocytes at both metaphase I and metaphase II stages. Morpholino-based depletion or genetic ablation of Pds5A all lead to defects in spindle organization, chromosome euploidy and meiotic progression in oocytes and thus compromising the female fertility. Mechanistically, Pds5A recruits deubiquitinase ubiquitin-specific protease 14 to the spindle apparatus for stabilization of kinesin family member 5B, regulating the spindle elongation. Collectively, our findings unveil that cohesin-associated protein Pds5A can be used as a spindle regulator during oocyte meiosis.
Figures


Similar articles
-
Manipulating Cohesin Levels in Live Mouse Oocytes.Methods Mol Biol. 2018;1818:113-128. doi: 10.1007/978-1-4939-8603-3_12. Methods Mol Biol. 2018. PMID: 29961260
-
Distinct roles of cohesin acetyltransferases Esco1 and Esco2 in porcine oocyte meiosis I.Cell Cycle. 2019 Oct;18(19):2481-2494. doi: 10.1080/15384101.2019.1651162. Epub 2019 Aug 6. Cell Cycle. 2019. PMID: 31387516 Free PMC article.
-
EZH2 is required for mouse oocyte meiotic maturation by interacting with and stabilizing spindle assembly checkpoint protein BubRI.Nucleic Acids Res. 2016 Sep 19;44(16):7659-72. doi: 10.1093/nar/gkw463. Epub 2016 May 25. Nucleic Acids Res. 2016. PMID: 27226494 Free PMC article.
-
Spindle formation, chromosome segregation and the spindle checkpoint in mammalian oocytes and susceptibility to meiotic error.Mutat Res. 2008 Mar 12;651(1-2):14-29. doi: 10.1016/j.mrgentox.2007.10.015. Epub 2007 Nov 9. Mutat Res. 2008. PMID: 18096427 Review.
-
The chromosomal basis of meiotic acentrosomal spindle assembly and function in oocytes.Chromosoma. 2017 Jun;126(3):351-364. doi: 10.1007/s00412-016-0618-1. Epub 2016 Nov 11. Chromosoma. 2017. PMID: 27837282 Free PMC article. Review.
References
-
- Xiong B., Gerton J. L., Regulators of the cohesin network. Annu. Rev. Biochem. 79, 131–153 (2010). - PubMed
-
- Ishiguro K.-i., Watanabe Y., Chromosome cohesion in mitosis and meiosis. J. Cell Sci. 120, 367–369 (2007). - PubMed
-
- Hirano T., Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69, 115–144 (2000). - PubMed
-
- Peters J.-M., Tedeschi A., Schmitz J., The cohesin complex and its roles in chromosome biology. Genes Dev. 22, 3089–3114 (2008). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

