Efficient and scalable construction of clinical variable networks for complex diseases with RAMEN
- PMID: 40215965
- PMCID: PMC12256955
- DOI: 10.1016/j.crmeth.2025.101022
Efficient and scalable construction of clinical variable networks for complex diseases with RAMEN
Abstract
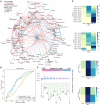
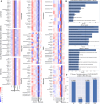
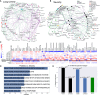
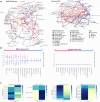
Understanding the interplay among clinical variables-such as demographics, symptoms, and laboratory results-and their relationships with disease outcomes is critical for advancing diagnostics and understanding mechanisms in complex diseases. Existing methods fail to capture indirect or directional relationships, while existing Bayesian network learning methods are computationally expensive and only infer general associations without focusing on disease outcomes. Here we introduce random walk- and genetic algorithm-based network inference (RAMEN), a method for Bayesian network inference that uses absorbing random walks to prioritize outcome-relevant variables and a genetic algorithm for efficient network refinement. Applied to COVID-19 (Biobanque québécoise de la COVID-19), intensive care unit (ICU) septicemia (MIMIC-III), and COPD (CanCOLD) datasets, RAMEN reconstructs networks linking clinical markers to disease outcomes, such as elevated lactate levels in ICU patients. RAMEN demonstrates advantages in computational efficiency and scalability compared to existing methods. By modeling outcome-specific relationships, RAMEN provides a robust tool for uncovering critical disease mechanisms, advancing diagnostics, and enabling personalized treatment strategies.
Keywords: Bayesian network inference; COVID-19; CP: Systems biology; absorbing random walk; chronic obstructive pulmonary disease; clinical variable networks; complex diseases; genetic algorithm; multi-omics; personalized medicine; septicemia.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Similar articles
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
ScITree: Scalable Bayesian inference of transmission tree from epidemiological and genomic data.PLoS Comput Biol. 2025 Jun 10;21(6):e1012657. doi: 10.1371/journal.pcbi.1012657. eCollection 2025 Jun. PLoS Comput Biol. 2025. PMID: 40493703 Free PMC article.
-
Automated monitoring compared to standard care for the early detection of sepsis in critically ill patients.Cochrane Database Syst Rev. 2018 Jun 25;6(6):CD012404. doi: 10.1002/14651858.CD012404.pub2. Cochrane Database Syst Rev. 2018. PMID: 29938790 Free PMC article.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
References
-
- Ding J., Hostallero D.E., El Khili M.R., Fonseca G.J., Milette S., Noorah N., Guay-Belzile M., Spicer J., Daneshtalab N., Sirois M., et al. A network-informed analysis of SARS-CoV-2 and hemophagocytic lymphohistiocytosis genes’ interactions points to neutrophil extracellular traps as mediators of thrombosis in COVID-19. PLoS Comput. Biol. 2021;17 - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

