Whole genome sequencing analysis of body mass index identifies novel African ancestry-specific risk allele
- PMID: 40216759
- PMCID: PMC11992084
- DOI: 10.1038/s41467-025-58420-2
Whole genome sequencing analysis of body mass index identifies novel African ancestry-specific risk allele
Abstract
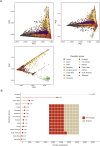
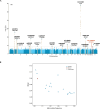
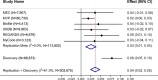
Obesity is a major public health crisis associated with high mortality rates. Previous genome-wide association studies (GWAS) investigating body mass index (BMI) have largely relied on imputed data from European individuals. This study leveraged whole-genome sequencing (WGS) data from 88,873 participants from the Trans-Omics for Precision Medicine (TOPMed) Program, of which 51% were of non-European population groups. We discovered 18 BMI-associated signals (P < 5 × 10-9), including two secondary signals. Notably, we identified and replicated a novel low-frequency single nucleotide polymorphism (SNP) in MTMR3 that was common in individuals of African descent. Using a diverse study population, we further identified two novel secondary signals in known BMI loci and pinpointed two likely causal variants in the POC5 and DMD loci. Our work demonstrates the benefits of combining WGS and diverse cohorts in expanding current catalog of variants and genes confer risk for obesity, bringing us one step closer to personalized medicine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: B.D.Ho. receives grant support from Bayer and has received an honorarium from AstraZeneca for an educational lecture. B.M.Ps. serve on the TOPMed Steering Committee. C.J.O. is employed by Novartis Institute of Biomedical Research, Cambridge, MA. D.L.D. received grants from Bayer and honoraria from Novartis. K.C.B. is an employee of Tempus. L.M.R. and S.S.R. are consultants for the TOPMed Administrative Coordinating Center (through Westat). U.P. was a consultant with AbbVie, and her husband is holding individual stocks for the following companies: BioNTech SE – ADR, Amazon, CureVac BV, NanoString Technologies, Google/Alphabet Inc Class C, NVIDIA Corp, Microsoft Corp. XLin is a consultant of AbbVie Pharmaceuticals and Verily Life Sciences. The remaining authors declare no competing interests.
Figures
Update of
-
WHOLE GENOME SEQUENCING ANALYSIS OF BODY MASS INDEX IDENTIFIES NOVEL AFRICAN ANCESTRY-SPECIFIC RISK ALLELE.medRxiv [Preprint]. 2023 Aug 22:2023.08.21.23293271. doi: 10.1101/2023.08.21.23293271. medRxiv. 2023. Update in: Nat Commun. 2025 Apr 11;16(1):3470. doi: 10.1038/s41467-025-58420-2. PMID: 37662265 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
- U01 HL054472/HL/NHLBI NIH HHS/United States
- R01 HL153460/HL/NHLBI NIH HHS/United States
- U19 CA203654/CA/NCI NIH HHS/United States
- R01 EB015611/EB/NIBIB NIH HHS/United States
- R01 DK075787/DK/NIDDK NIH HHS/United States
- F32 HL085989/HL/NHLBI NIH HHS/United States
- U01 HL072524/HL/NHLBI NIH HHS/United States
- I01 BX003340/BX/BLRD VA/United States
- P20 GM109036/GM/NIGMS NIH HHS/United States
- P01 CA134294/CA/NCI NIH HHS/United States
- U01 HG009088/HG/NHGRI NIH HHS/United States
- T32 HL007055/HL/NHLBI NIH HHS/United States
- R01 DK122503/DK/NIDDK NIH HHS/United States
- R35 CA197449/CA/NCI NIH HHS/United States
- R01 HL104135/HL/NHLBI NIH HHS/United States
- I01 BX004821/BX/BLRD VA/United States
- R01 HL133040/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R01 AR048797/AR/NIAMS NIH HHS/United States
- U01 HG007416/HG/NHGRI NIH HHS/United States
- U01 HL054509/HL/NHLBI NIH HHS/United States
- R01 DK 122503/U.S. Department of Health & Human Services | NIH | Office of Extramural Research, National Institutes of Health (OER)
- U01 HL120393/HL/NHLBI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- R01 HL113338/HL/NHLBI NIH HHS/United States
- R01 DK135938/DK/NIDDK NIH HHS/United States
- R01 AG058921/AG/NIA NIH HHS/United States
- R01 HL142302/HL/NHLBI NIH HHS/United States
- R01 DK110113/DK/NIDDK NIH HHS/United States
- R01 NS058700/NS/NINDS NIH HHS/United States
- R01 DK107786/DK/NIDDK NIH HHS/United States
- R01 HL119443/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 AI114555/AI/NIAID NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- T32 HL129982/HL/NHLBI NIH HHS/United States
- R01 HL067348/HL/NHLBI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 HL172803/HL/NHLBI NIH HHS/United States
- R01 HL104608/HL/NHLBI NIH HHS/United States
- R01 AI132476/AI/NIAID NIH HHS/United States
- K08 HL136928/HL/NHLBI NIH HHS/United States
- M01 RR007122/RR/NCRR NIH HHS/United States
- KL2 TR002490/TR/NCATS NIH HHS/United States
- R01 HL142825/HL/NHLBI NIH HHS/United States
- R01 DK124097/DK/NIDDK NIH HHS/United States
- R01 HL093093/HL/NHLBI NIH HHS/United States
- I01 BX003362/BX/BLRD VA/United States
- R01 HL068959/HL/NHLBI NIH HHS/United States
- U01 HL072507/HL/NHLBI NIH HHS/United States
- R01 DK071891/DK/NIDDK NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- R01 HG010297/HG/NHGRI NIH HHS/United States
- P30 CA071789/CA/NCI NIH HHS/United States
- U01 HL054495/HL/NHLBI NIH HHS/United States
- R01 HL055673/HL/NHLBI NIH HHS/United States
- R01 HL092301/HL/NHLBI NIH HHS/United States
- U01 HL054473/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

