Natural killer cells' functional impairment drives the immune escape of pre-malignant clones in early-stage myelodysplastic syndromes
- PMID: 40216768
- PMCID: PMC11992119
- DOI: 10.1038/s41467-025-58662-0
Natural killer cells' functional impairment drives the immune escape of pre-malignant clones in early-stage myelodysplastic syndromes
Abstract
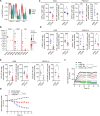
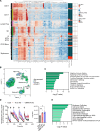
Dissecting the preneoplastic disease states' biological mechanisms that precede tumorigenesis can lead to interventions that can slow down disease progression and/or mitigate disease-related comorbidities. Myelodysplastic syndromes (MDS) cannot be cured by currently available pharmacological therapies, which fail to eradicate aberrant hematopoietic stem cells (HSCs), most of which are mutated by the time of diagnosis. Here, we sought to elucidate how MDS HSCs evade immune surveillance and expand in patients with clonal cytopenias of undetermined significance (CCUS), the pre-malignant stage of MDS. We used multi-omic single-cell approaches and functional in vitro studies to show that immune escape at disease initiation is mainly mediated by mutant, dysfunctional natural killer (NK) cells with impaired cytotoxic capability against cancer cells. Preclinical in vivo studies demonstrated that injecting NK cells from healthy donors efficiently depleted CCUS mutant cells while allowing normal cells to regenerate hematopoiesis. Our findings suggest that early intervention with adoptive cell therapy can prevent or delay the development of MDS.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All authors declare no competing interests relative to this work.
Figures


References
-
- de Visser, K. E. & Joyce, J. A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell41, 374–403 (2023). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

