Autophagy is induced during plant grafting to promote wound healing
- PMID: 40216774
- PMCID: PMC11992148
- DOI: 10.1038/s41467-025-58519-6
Autophagy is induced during plant grafting to promote wound healing
Abstract
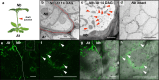
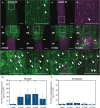
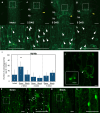
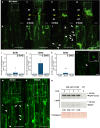
Grafting is an agricultural technique that joins tissues from different plants to obtain useful rootstock traits. However, cellular processes involved in joint tissue repair remain poorly understood. We analyzed Nicotiana benthamiana (Nb) and Arabidopsis thaliana (At) interfamily heterografting as a high-stress model and At homografting as a low-stress model. Transmission electron micrographs reveal the formation of autophagic structures in cells near the graft boundary over a long period in Nb/At interfamily grafts and in a short period of a few days in At homografts. Using a GFP-ATG8 marker line, the autophagosomes were observed in the cells near the graft boundary, especially on the scion side, where nutrient depletion occurred. Grafting of At autophagy-defective mutants decreases grafting success rates and post-grafting growth. NbATG5 knockdown suppresses graft establishment in Nb/At interfamily heterografts. Moreover, At autophagy-defective mutants show reduced callus formation directed to wounds under the nutrient-deficient conditions. These results suggest that autophagy is induced during grafting, promoting callus formation and contributing to tissue connectivity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Davies, Jr. F. T., Geneve, R. L. & Wilson, S. B. Hartmann & Kester’s Plant Propagation: Principles and Practices, 9thedn. 490–542 (Pearson, NY,2018).
-
- Gaut, B. S., Miller, A. J. & Seymour, D. K. Living with two genomes: grafting and its implications for plant genome-to-genome interactions, phenotypic variation, and evolution. Annu. Rev. Genet.53, 51–5.21 (2019). - PubMed
-
- Thomas, H. R. & Frank, M. H. Connecting the pieces: uncovering the molecular basis for long-distance communication through plant grafting. N. Phytol.223, 582–589 (2019). - PubMed
-
- Notaguchi, M. et al. Cell-cell adhesion in plant grafting is facilitated by β-1,4-glucanases. Science369, 698–702 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- 18KT0040/MEXT | Japan Society for the Promotion of Science (JSPS)
- 19H05361/MEXT | Japan Society for the Promotion of Science (JSPS)
- 21H00368/MEXT | Japan Society for the Promotion of Science (JSPS)
- 21H05657/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22K06181/MEXT | Japan Society for the Promotion of Science (JSPS)
- 19H05713/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22H04926/MEXT | Japan Society for the Promotion of Science (JSPS)
- 24H02142/MEXT | Japan Society for the Promotion of Science (JSPS)
- JPMJTR194G/MEXT | Japan Science and Technology Agency (JST)
- JPNP20004/New Energy and Industrial Technology Development Organization (NEDO)

