A role for astrocytic miR-129-5p in frontotemporal dementia
- PMID: 40216778
- PMCID: PMC11992244
- DOI: 10.1038/s41398-025-03338-y
A role for astrocytic miR-129-5p in frontotemporal dementia
Abstract
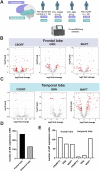
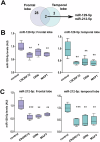
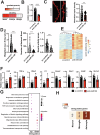
Frontotemporal dementia is a debilitating neurodegenerative disorder characterized by frontal and temporal lobe degeneration, resulting in behavioral changes, language difficulties, and cognitive decline. In this study, smallRNA sequencing was conducted on postmortem brain tissues obtained from the frontal and temporal of FTD patients with GRN, MAPT, or C9ORF72 mutations. Our analysis identified miR-129-5p as consistently deregulated across all analyzed mutation conditions and brain regions. Functional investigations in in-vitro models revealed a novel role of miR-129-5p in astrocytes, where its loss led to neuroinflammation and impaired neuronal support functions, including reduced glutamate uptake. Depletion of miR-129-5p in astrocytes also resulted in the loss of neuronal spines and altered neuronal network activity in a cell culture system. These findings highlight miR-129-5p as a potential therapeutic target in neurodegenerative diseases and also sheds light on the role of astrocytes in Frontotemporal dementia pathogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no conflict of interest.
Figures



References
-
- Younes K, Miller BL. Frontotemporal dementia: neuropathology, genetics, neuroimaging, and treatments. Psychiatr Clin North Am. 2020;43:331–44. - PubMed
-
- Ratnavalli E, Brayne C, Dawson K, Hodges JR. The prevalence of frontotemporal dementia. Neurology. 2002;58:1615–21. - PubMed
-
- Mackenzie IR, Neumann M. Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J Neurochem. 2016;138:54–57. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

