Myeloid lineage C3 induces reactive gliosis and neuronal stress during CNS inflammation
- PMID: 40216817
- PMCID: PMC11992029
- DOI: 10.1038/s41467-025-58708-3
Myeloid lineage C3 induces reactive gliosis and neuronal stress during CNS inflammation
Abstract
Complement component C3 mediates pathology in CNS neurodegenerative diseases. Here we use scRNAseq of sorted C3-reporter positive cells from mouse brain and optic nerve to characterize C3 producing glia in experimental autoimmune encephalomyelitis (EAE), a model in which peripheral immune cells infiltrate the CNS, causing reactive gliosis and neuro-axonal pathology. We find that C3 expression in the early inflammatory stage of EAE defines disease-associated glial subtypes characterized by increased expression of genes associated with mTOR activation and cell metabolism. This pro-inflammatory subtype is abrogated with genetic C3 depletion, a finding confirmed with proteomic analyses. In addition, early optic nerve axonal injury and retinal ganglion cell oxidative stress, but not loss of post-synaptic density protein 95, are ameliorated by selective deletion of C3 in myeloid cells. These data suggest that in addition to C3b opsonization of post synaptic proteins leading to neuronal demise, C3 activation is a contributor to reactive glia in the optic nerve.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




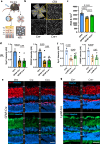
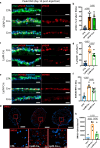
References
-
- Qian, K. et al. Revisiting the critical roles of reactive astrocytes in neurodegeneration. Mol. Psychiatry28, 2697–2706 (2023). - PubMed
MeSH terms
Substances
Grants and funding
- RG-1907- 992 34756/National Multiple Sclerosis Society (National MS Society)
- T32 GM136577/GM/NIGMS NIH HHS/United States
- R01NS041435/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- S10 OD021844/OD/NIH HHS/United States
- High-End Instrumentation grant (S10OD021844)/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

