Mutualism breakdown underpins evolutionary rescue in an obligate cross-feeding bacterial consortium
- PMID: 40216843
- PMCID: PMC11992082
- DOI: 10.1038/s41467-025-58742-1
Mutualism breakdown underpins evolutionary rescue in an obligate cross-feeding bacterial consortium
Abstract
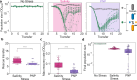
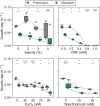
Populations facing lethal environmental change can escape extinction through rapid genetic adaptation, a process known as evolutionary rescue. Despite extensive study, evolutionary rescue is largely unexplored in mutualistic communities, where it is likely constrained by the less adaptable partner. Here, we explored empirically the likelihood, population dynamics, and genetic mechanisms underpinning evolutionary rescue in an obligate mutualism involving cross-feeding of amino acids between auxotrophic Escherichia coli strains. We found that over 80% of populations overcame a severe decline when exposed to two distinct types of abrupt, lethal stress. Of note, in all cases only one of the strains survived by metabolically bypassing the auxotrophy. Crucially, the mutualistic consortium exhibited greater sensitivity to both stressors than a prototrophic control strain, such that reversion to autonomy was sufficient to alleviate stress below lethal levels. This sensitivity was common across other stresses, suggesting it may be a general feature of amino acid-dependent obligate mutualisms. Our results reveal that evolutionary rescue may depend critically on the specific genetic and physiological details of the interacting partners, adding rich layers of complexity to the endeavor of predicting the fate of microbial communities facing intense environmental deterioration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declared no competing interests.
Figures



Similar articles
-
Obligate mutualistic cooperation limits evolvability.Nat Commun. 2022 Jan 17;13(1):337. doi: 10.1038/s41467-021-27630-9. Nat Commun. 2022. PMID: 35039522 Free PMC article.
-
Exometabolomics Assisted Design and Validation of Synthetic Obligate Mutualism.ACS Synth Biol. 2016 Jul 15;5(7):569-76. doi: 10.1021/acssynbio.5b00236. Epub 2016 Feb 17. ACS Synth Biol. 2016. PMID: 26885935
-
Mutualisms in a changing world: an evolutionary perspective.Ecol Lett. 2010 Dec;13(12):1459-74. doi: 10.1111/j.1461-0248.2010.01538.x. Epub 2010 Oct 19. Ecol Lett. 2010. PMID: 20955506
-
The evolution of plant-insect mutualisms.New Phytol. 2006;172(3):412-28. doi: 10.1111/j.1469-8137.2006.01864.x. New Phytol. 2006. PMID: 17083673 Review.
-
Evolution on the bright side of life: microorganisms and the evolution of mutualism.Ann N Y Acad Sci. 2018 Jun;1422(1):88-103. doi: 10.1111/nyas.13515. Epub 2017 Nov 30. Ann N Y Acad Sci. 2018. PMID: 29194650 Review.
Cited by
-
Strengthen or Weaken: Evolutionary Directions of Cross-Feeding After Formation.Environ Microbiol Rep. 2025 Aug;17(4):e70175. doi: 10.1111/1758-2229.70175. Environ Microbiol Rep. 2025. PMID: 40784676 Free PMC article. Review.
References
-
- Bell, G. Evolutionary rescue. Annu. Rev. Ecol. Evol. Syst.48, 605–627 (2017).
-
- Diniz-Filho, J. A. F. & Bini, L. M. Will life find a way out? evolutionary rescue and Darwinian adaptation to climate change. Perspect. Ecol. Conserv.17, 117–121 (2019).
-
- Carlson, S. M., Cunningham, C. J. & Westley, P. A. H. Evolutionary rescue in a changing world. Trends Ecol. Evol.29, 521–530 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

