In silico pan-cancer analysis of VRAC subunits and their prognostic roles in human cancers
- PMID: 40216864
- PMCID: PMC11992229
- DOI: 10.1038/s41598-025-97078-0
In silico pan-cancer analysis of VRAC subunits and their prognostic roles in human cancers
Abstract
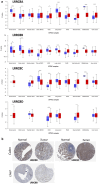
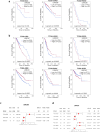
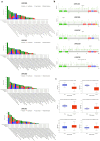
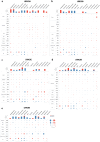
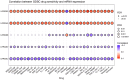
The study focuses on the VRAC channel and its significant roles in cancer development. It addresses a research gap by conducting a pan-cancer analysis with multi-omics bioinformatics tools, integrating data from the Human Protein Atlas (HPA) and Genotype-Tissue Expression (GTEx) datasets to examine mRNA expression patterns of its Leucine Rich Repeat Containing 8 (LRRC8) subunits in various tissues and cancers. The study links variations in LRRC8s expression with patient outcomes and includes analyses of DNA and RNA methylation. The study reveals significant correlations between LRRC8s expression and immune cell infiltration, as well as a positive association with cancer-associated fibroblasts and key immune regulators such as major histocompatibility complex (MHCs) and chemokines. Furthermore, the research suggests that LRRC8s are involved in cancer-signalling pathways, which may offer new therapeutic targets. Additionally, a drug sensitivity analysis shows that LRRC8 subunits affect drug responses differently, supporting the use of personalized therapeutic strategies. In conclusion, the study emphasizes the significance of VRAC subunits in cancer biology and suggests their potential as biomarkers and targets in cancer immunotherapy and personalized medicine.
Keywords: Immuno-oncology; LRRC8 subunits; Prognosis; VRAC.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures







Similar articles
-
Pan-cancer analysis reveals correlation between RAB3B expression and tumor heterogeneity, immune microenvironment, and prognosis in multiple cancers.Sci Rep. 2024 Apr 30;14(1):9881. doi: 10.1038/s41598-024-60581-x. Sci Rep. 2024. PMID: 38688977 Free PMC article.
-
Multiomic characterization, immunological and prognostic potential of SMAD3 in pan-cancer and validation in LIHC.Sci Rep. 2025 Jan 3;15(1):657. doi: 10.1038/s41598-024-84553-3. Sci Rep. 2025. PMID: 39753728 Free PMC article.
-
Comprehensive pan-cancer analysis reveals prognostic implications of TMEM92 in the tumor immune microenvironment.Clin Transl Oncol. 2024 Oct;26(10):2701-2717. doi: 10.1007/s12094-024-03477-6. Epub 2024 Apr 20. Clin Transl Oncol. 2024. PMID: 38642258
-
Volume-regulated anion channel as a novel cancer therapeutic target.Int J Biol Macromol. 2020 Sep 15;159:570-576. doi: 10.1016/j.ijbiomac.2020.05.137. Epub 2020 May 20. Int J Biol Macromol. 2020. PMID: 32442571 Review.
-
The volume-regulated anion channel is formed by LRRC8 heteromers – molecular identification and roles in membrane transport and physiology.Biol Chem. 2015 Sep;396(9-10):975-90. doi: 10.1515/hsz-2015-0127. Biol Chem. 2015. PMID: 25868000 Review.
References
-
- Orsolic, I., Carrier, A. & Esteller, M. Genetic and epigenetic defects of the RNA modification machinery in cancer. Trends Genet.39(1), 74–88 (2023). - PubMed
-
- Ilango, S. et al. Epigenetic alterations in cancer. Front. Biosci. (Landmark Ed.)25(6), 1058–1109 (2020). - PubMed
-
- Yu, B., Shao, S. & Ma, W. Frontiers in pancreatic cancer on biomarkers, microenvironment, and immunotherapy. Cancer Lett.610, 217350 (2025). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

